Researchers have utilized a powerful new technique called spatial transcriptomics to gain a deeper understanding of chronic hepatitis B (CHB) infection, a serious liver disease affecting millions of people worldwide. This study sheds light on the intricate interplay between the virus and the immune system within the liver, paving the way for developing novel therapeutic strategies.
CHB is a global health burden with limited treatment options. Current therapies can suppress viral replication but do not eliminate the virus. This leaves patients at risk for complications like cirrhosis and liver cancer. The complexities of the immune response in CHB further hinder the development of effective cures.
READ MORE: Researchers may have found an Achilles heel for Hepatitis B
“Understanding the spatial distribution of infected cells and immune activity within the liver tissue is crucial for developing targeted therapies for CHB,” explains Dr. Amy Cross, lead author of the study. “Spatial transcriptomics allows us to examine gene expression patterns at a microscopic level, revealing previously hidden details about the disease process.”
Analyzing liver biopsies
The researchers employed spatial transcriptomics to analyze liver biopsies from patients with CHB, including those with hepatitis D virus (HDV) or human immunodeficiency virus (HIV) co-infection. Their findings revealed the existence of distinct gene expression signatures associated with the presence of infected cells and immune cell infiltration. They also found that all patients had shared features, such as genes involved in B cell signalling and cytotoxicity.
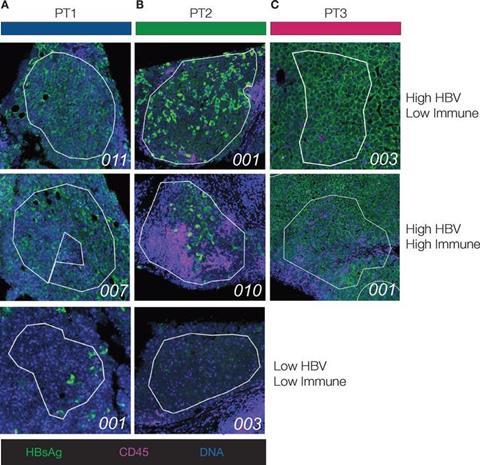
The researchers also found unique signatures in patients with co-infections, suggesting potential disease progression mechanisms. The differences in immune cell composition between patients highlighted the variability of the immune response in CHB.
“This study demonstrates the immense potential of spatial transcriptomics for unravelling the complexities of CHB infection,” say Dr Dimitra Peppa and Dr Upkar Gill, senior authors of the study. “By pinpointing specific pathways and immune cell populations involved in the disease process, we can identify new therapeutic targets and develop personalized treatment strategies.”
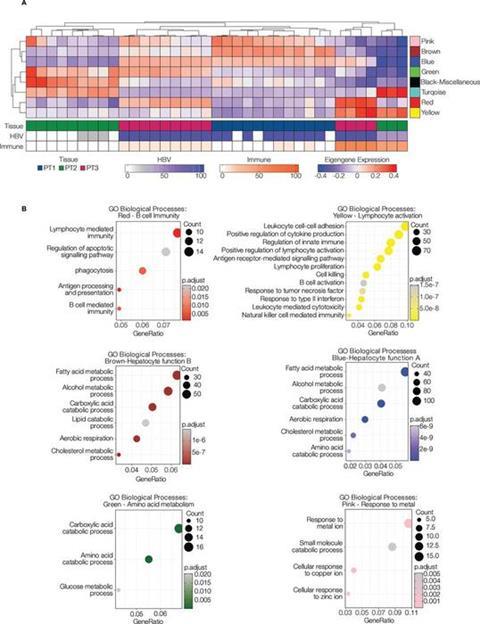
This research paves the way for further investigations using spatial transcriptomics to explore the intricate mechanisms of CHB infection and co-infections. By gaining a deeper understanding of the disease at the cellular and molecular level, researchers can work towards more effective treatments and, ultimately, a cure for CHB.






No comments yet