With antibiotic resistance a growing problem, University of Virginia School of Medicine researchers have developed cutting-edge computer models that could give the disease-fighting drugs a laser-like precision to target only specific bacteria in specific parts of the body. Their findings are published in the scientific journal PLOS Biology.
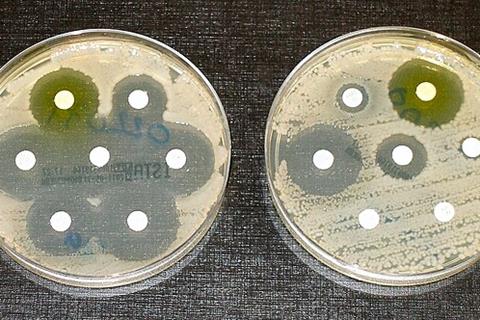
As it stands, antibiotics kill bacteria indiscriminately. Because the drugs are used so widely, increasing numbers of dangerous bugs are growing resistant, threatening one of modern medicine’s most important weapons against disease.
Alternative way to tackle antibiotic resistance
UVA’s new approach, on the other hand, would dramatically limit how often bacteria are exposed to antibiotics, reducing the chance they could become resistant to antibiotics. Further, the approach would represent a significant step forward for precision medicine, allowing doctors to better tailor treatments to individual patients’ needs. Instead of taking an antibiotic that kills bacteria regardless of whether helpful or harmful, patients could be given antibiotics that target specific bacteria causing a specific problem in a specific area of the body.
READ MORE: Computer model of influenza virus shows universal vaccine promise
READ MORE: Researchers use AI to help detect antibiotic resistance
“Many biomedical challenges are incredibly complex, and computer models are emerging as a powerful tool for tackling such problems,” said researcher Jason Papin, PhD, of UVA’s Department of Biomedical Engineering. “We’re hopeful that these computer models of the molecular networks in bacteria will help us develop new strategies to treat infections.”
More targeted antibiotics
UVA’s new approach was made possible by a herculean effort by Papin, PhD student Emma Glass and their collaborators. Working with Andrew Warren, PhD, of UVA’s Biocomplexity Institute, the researchers in Papin’s lab developed sophisticated computer models of every human bacterial pathogen with sufficient genetic information available.
Glass then analyzed all those models and identified shared traits among the bacteria. This analysis yielded the discovery that bacteria in certain parts of the body, such as the stomach, tended to share metabolic properties. Basically, where they live shapes how they function.
“Using our computer models we found that the bacteria living in the stomach had unique properties,” Glass said. “These properties can be used to guide design of targeted antibiotics, which could hopefully one day slow the emergence of resistant infections.”
Future of computer modelling in biomedical research
The shared similarities among the microbes in different locales could be the Achilles’ heel for harmful bacteria in our bodies. With further research, doctors may be able to target specific types of bacteria in specific areas, reducing the need for broad-spectrum antibiotics.
Putting their computer-modeling approach to the test, Papin and his team have already found that they could inhibit the growth of harmful stomach bugs in lab experiments. That’s a promising sign for the future potential of their computer-modelling approach.
“We still have much to do to test these ideas for other bacteria and types of infections,” Papin said. “but this work shows the incredible promise of data science and computer modelling for tackling some of the most important problems in biomedical research.
Topics
- Andrew Warren
- Antibiotic resistance
- Antibiotics
- Antimicrobial Resistance
- Bacteria
- Computational Biology
- computer-modeling
- Early Career Research
- Emma Glass
- Healthy Land
- Innovation News
- Jason Papin
- One Health
- University of Virginia School of Medicine
- University of Virginia School of Medicine Biocomplexity Institute
- USA & Canada
- Young Innovators







No comments yet