Genome editing holds great promise for the molecular breeding of plants, yet its application is hindered by the shortage of simple and effective means of delivering genome editing reagents into plants.

Conventional plant transformation-based methods for the delivery of genome editing reagents into plants often involve prolonged tissue culture, a labor-intensive and technically challenging process for many elite crop cultivars. This bottleneck prevents the genomes from many elite crop cultivars from being edited in a cost-effective manner.
Genome editing strategies based on homology-directed repair (HDR) are highly versatile, but often depend on a large amount of donor DNA delivered, which can be difficult to achieve with many conventional methods. Both RNA viruses and DNA viruses have been employed to overcome these technical challenges, in a study from Nanjing Agricultural University published in aBIOTECH.
Harnessing RNA viruses
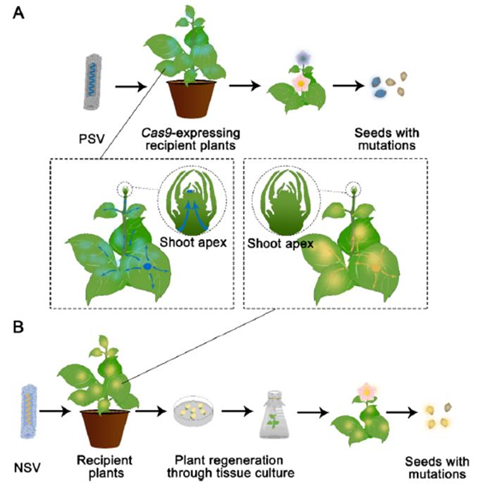
RNA viruses have been harnessed for the delivery of the core components involved in genome editing in plants. The limitation in the capacity of positive-strand RNA viruses (PSVs) is compensated by their ability to invade germline cells in planta and induce heritable edits. Accordingly, PSV-based delivery strategies have been mostly applied to introduce guide RNA molecules into plant hosts that express the matching Cas nuclease gene, which often exceeds the size limit that PSVs can accommodate.
On the contrary, negative-strand RNA viruses (NSVs) can accommodate longer DNA fragments, such as one encoding the entire sequence-specific nuclease machinery, but rarely enters the germline cells. As a result, NSVs-based delivery strategies often entail a subsequent plant regeneration process if heritable edits are desired.
Precision edits achieved via HDR usually involves an artificially supplied donor DNA as the repair template, which needs to be delivered at a high copy number to be effective. Geminiviruses comprise a family of plant DNA viruses whose genomes can replicate to a very high copy number in plant cells. This feature makes them ideal vectors for the delivery of repair donors. Geminiviral replicons have been successfully utilized to generate specific editing outcomes in wheat, tomatoes, and tobacco etc.
Trade-off
A general trade-off between cargo capacity and vector mobility exists for currently available viral vectors. PSVs are promising tools for tissue culture-free gene editing, but they rely on an existing Cas-expressing line due to the limited capacity of the viral vector.
NSV-based vectors can accommodate the entire CRISPR/Cas machinery, and thus can be used for genome editing in a transgene-free context, but often rely on a subsequence tissue culture process to recover plants carrying heritable edits. Similarly, GVRs are modified into replicon vectors with no infectivity and minimal mobility to make room for extra nucleotide sequences.
It is desirable to develop viral vector systems with not only the ability to perform cargo delivery into germline cells, in planta, but also sufficient capacity for the complete CRISPR/Cas components. Meanwhile, more compact sequence-specific nucleases are strong candidates to be delivered using virus-based systems.
Besides, it is worth exploring new components to be fused with the delivered cargos to enhance the systemic movement of the genome editing reagents to achieve germline edits. Furthermore, broad-spectrum viral delivery systems are required for application in a broader range of crop species. Biosafety and risk assessment of the application viral vectors are also worth additional investigation to reduce unintended burden on humans and the ecosystem.
Potential solutions
In summary, the use of viral vectors to deliver genome editing components offers potential solutions to many current technical bottlenecks involved in genome editing in plants. More efficient delivery methods capable of generating heritable edits in a simple manner may be established in the future through the exploitation of novel viral species and engineered existing viruses for improved performance.







No comments yet