The bacterium Deinococcus radiodurans can survive a range of harsh environments and is resistant to very high levels of ionizing and UV radiation, dehydration, cold, vacuum, and even acid, with potential to act as a bacterial chassis (a bacterium that carries and supports the genetic components necessary for a particular experiment or application) in synthetic biology.

For instance, it can operate as a biological factory in the industrial production of valuable compounds, help in nuclear waste or soil treatment, and assist in the remediation of oil spills. However, genetic engineering tools specific to D. radiodurans are required to realize the bacterium’s potential in human applications.
In a study published in BioDesign Research, researchers from the University of Western Ontario discuss their design of a new method for performing seamless gene deletions in D. radiodurans.
This innovation, named SLICER, may make the genetic engineering of D. radiodurans significantly easier since the bacterium’s genome has historically been hard to manipulate.
”Many microorganisms possess immune mechanisms called restriction-modification (R-M) systems that protect against foreign DNA molecules. Previous studies have identified some R-M systems in D. radiodurans, which prevent efficient genetic manipulation of the bacterium,” explains Professor Bogumil Karas, the corresponding author of the study.
Recombination mechanism
To develop the SLICER method (an abbreviation for seamless loss of integrated cassettes using endonuclease cleavage and recombination), the researchers exploited a recombination mechanism in D. radiodurans responsible for the bacterium’s high resistance to radiation and other stressors. The mechanism is called “homologous recombination”, a process by which D. radiodurans swaps corresponding parts of its two chromosomes to repair damaged DNA.
The SLICER method involves three steps. In the first step, a strand of DNA called a seamless deletion (SD) cassette is inserted into the bacteria. The strand of DNA is flanked by an identical pair of DNA regions, forming the gene of interest (GOI), which needs to be deleted.
The bacteria’s homologous recombination mechanism then replaces the GOI with the SD cassette in some, but not all, bacteria. The SD cassette also contains genes that provide the bacteria resistance to certain antibiotics. Bacteria in which the SD cassette has successfully replaced the GOI survive and produce colonies in a growth medium containing the antibiotic.
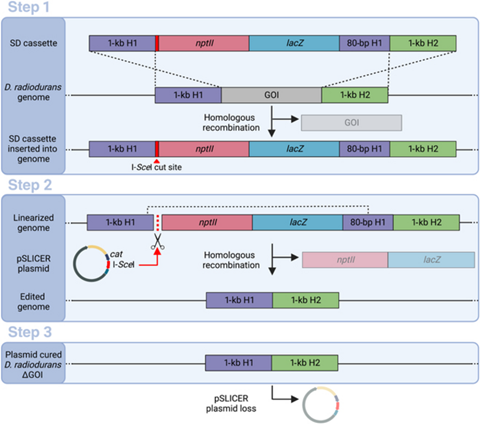
In the second step, after the bacteria with the SD cassette has been isolated using antibiotic selection, a circular DNA strand called “pSLICER plasmid” is introduced into the cell. The plasmid then produces an endonuclease enzyme that makes a cut in the SD cassette, prompting the bacteria to remove it using homologous recombination. In the third step, the pSLICER plasmid is eliminated, resulting in a bacterial strain lacking GOI.
The researchers selectively removed five of the six known R-M systems in D. radiodurans to demonstrate the effectiveness of the SLICER method. Bacterial growth and resistance to foreign DNA was unaffected by this.
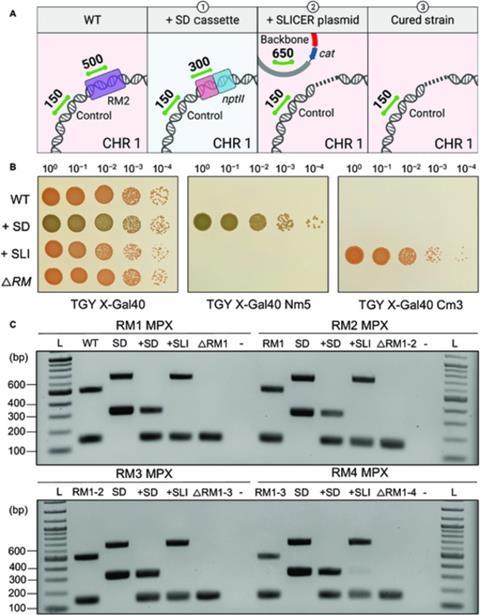
Professor Karas concludes: ”While transformation was not significantly improved in our final strain, the SLICER method was demonstrated as an efficient method for engineering D. radiodurans. It could enable the deletion of multiple genes of interest (GOI) and ultimately lead to the further development of laboratory or industrial strains, with multiple applications.”
Topics
- Bacteria
- bacterial chassis
- Bogumil Karas
- Climate Action
- Deinococcus radiodurans
- Future Technologies
- Future Technologies
- Healthy Land
- Industrial Microbiology
- Microbial Genomics
- One Health
- pSLICER plasmid
- Research News
- restriction-modification systems
- SLICER
- Synthetic Biology
- University of Western Ontario
- USA & Canada







No comments yet