Marine algae Prymnesium parvum use massive enzymes dubbed PKZILLAs – some of the largest proteins ever to be identified in nature – to make large and complex prymnesin neurotoxins responsible for mass fish kills during harmful algal blooms worldwide, researchers report.
While seeking to unravel how marine algae create their chemically complex toxins, scientists at UC San Diego’s Scripps Institution of Oceanography discovered the largest protein yet identified in biology. Uncovering the biological machinery the algae evolved to make its intricate toxin also revealed previously unknown strategies for assembling chemicals, which could unlock the development of new medicines and materials.
“The discovery and initial characterization of the prymnesin PKZILLA gigasynthases now elucidates the long-standing question about how microalgae biosynthesize their giant polyketide polyether molecules,” write the authors.
READ MORE: Researchers discover genetic collaboration in harmful algae
READ MORE: Climate change is increasing risk of high toxin concentrations in Northern US lakes
It also expands expectations of genetic and enzymatic size limits in biology.
Exotic molecules
Many marine microbes produce exotic organic molecules with varied biological functions. Some microalgae, like P. parvum, are known for producing some of the largest nonpolymeric carbon chain molecules in nature, including polyketide polyether biotoxins.
During harmful algal blooms, neurotoxic prymnesins compounds are notorious for causing environmental damage, including massive environmental fish kills. However, despite decades of extensive research, how these microalgae produce such large and complex compounds is poorly understood.
Massive enzymes
Using a customized gene annotation strategy, Timothy Fallon and colleagues discovered genes in P. parvum, which they named PKZILLAs (PKZILLA-1 and PKZILLA-2), that are involved in the production of polyketide synthase (PKS) enzymes.
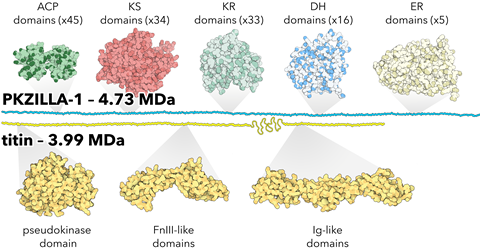
Notably, Fallon et al. found that these enzymes were massive, with PKZILLA-1 being one of the largest proteins ever identified at 4.7 megadaltons and containing 140 enzyme domains. Although slightly smaller, PKZILLA-2 is 3.2 megadaltons with 99 enzyme domains.
According to the findings, these massive PKS gigasynthases are responsible for the biosynthesis of the 90-carbon backbone of prymnesin toxins. The authors also characterized a variant, PKZILLA-B1, which produces a shorter version of these toxins.
Researchers found the protein while studying how a type of algae called Prymnesium parvum makes its toxin, which is responsible for massive fish kills.
Mount Everest
“This is the Mount Everest of proteins,” said Bradley Moore, a marine chemist with joint appointments at Scripps Oceanography and Skaggs School of Pharmacy and Pharmaceutical Sciences and senior author of a new study detailing the findings. “This expands our sense of what biology is capable of.”
PKZILLA-1 is 25% larger than titin, the previous record holder, which is found in human muscles and can reach 1 micron in length (0.0001 centimeter or 0.00004 inch).
Published today in Science and funded by the National Institutes of Health and the National Science Foundation, the study shows that this giant protein and another super-sized but not record-breaking protein – PKZILLA-2 – are key to producing prymnesin – the big, complex molecule that is the algae’s toxin. In addition to identifying the massive proteins behind prymnesin, the study also uncovered unusually large genes that provide Prymnesium parvum with the blueprint for making the proteins.
Monitoring efforts
Finding the genes that undergird the production of the prymnesin toxin could improve monitoring efforts for harmful algal blooms from this species by facilitating water testing that looks for the genes rather than the toxins themselves.
“Monitoring for the genes instead of the toxin could allow us to catch blooms before they start instead of only being able to identify them once the toxins are circulating,” said Timothy Fallon, a postdoctoral researcher in Moore’s lab at Scripps and co-first author of the paper.
Discovering the PKZILLA-1 and PKZILLA-2 proteins also lays bare the alga’s elaborate cellular assembly line for building the toxins, which have unique and complex chemical structures. This improved understanding of how these toxins are made could prove useful for scientists trying to synthesize new compounds for medical or industrial applications.
“Understanding how nature has evolved its chemical wizardry gives us as scientific practitioners the ability to apply those insights to creating useful products, whether it’s a new anti-cancer drug or a new fabric,” said Moore.
Golden algae
Prymnesium parvum, commonly known as golden algae, is an aquatic single-celled organism found all over the world in both fresh and saltwater. Blooms of golden algae are associated with fish die offs due to its toxin prymnesin, which damages the gills of fish and other water breathing animals. In 2022, a golden algae bloom killed 500-1,000 tons of fish in the Oder River adjoining Poland and Germany. The microorganism can cause havoc in aquaculture systems in places ranging from Texas to Scandinavia.
Prymnesin belongs to a group of toxins called polyketide polyethers that includes brevetoxin B, a major red tide toxin that regularly impacts Florida, and ciguatoxin, which contaminates reef fish across the South Pacific and Caribbean. These toxins are among the largest and most intricate chemicals in all of biology, and researchers have struggled for decades to figure out exactly how microorganisms produce such large, complex molecules.
Toxin mystery
Beginning in 2019, Moore, Fallon and Vikram Shende, a postdoctoral researcher in Moore’s lab at Scripps and co-first author of the paper, began trying to figure out how golden algae make their toxin prymnesin on a biochemical and genetic level.
The study authors began by sequencing the golden alga’s genome and looking for the genes involved in producing prymnesin. Traditional methods of searching the genome didn’t yield results, so the team pivoted to alternate methods of genetic sleuthing that were more adept at finding super long genes.
“We were able to locate the genes, and it turned out that to make giant toxic molecules this alga uses giant genes,” said Shende.
With the PKZILLA-1 and PKZILLA-2 genes located, the team needed to investigate what the genes made to tie them to the production of the toxin. Fallon said the team was able to read the genes’ coding regions like sheet music and translate them into the sequence of amino acids that formed the protein.
Record-breaking mass
When the researchers completed this assembly of the PKZILLA proteins they were astonished at their size. The PKZILLA-1 protein tallied a record-breaking mass of 4.7 megadaltons, while PKZILLA-2 was also extremely large at 3.2 megadaltons. Titin, the previous record-holder, can be up to 3.7 megadaltons – about 90-times larger than a typical protein
.
After additional tests showed that golden algae actually produce these giant proteins in life, the team sought to find out if the proteins were involved in making the toxin prymnesin. The PKZILLA proteins are technically enzymes, meaning they kick off chemical reactions, and the team played out the lengthy sequence of 239 chemical reactions entailed by the two enzymes with pens and notepads.
“The end result matched perfectly with the structure of prymnesin,” said Shende.
Cascade of reactions
Following the cascade of reactions that golden algae uses to make its toxin revealed previously unknown strategies for making chemicals in nature, said Moore. “The hope is that we can use this knowledge of how nature makes these complex chemicals to open up new chemical possibilities in the lab for the medicines and materials of tomorrow,” he added.
Finding the genes behind the prymnesin toxin could allow for more cost effective monitoring for golden algae blooms. Such monitoring could use tests to detect the PKZILLA genes in the environment akin to the PCR tests that became familiar during the COVID-19 pandemic. Improved monitoring could boost preparedness and allow for more detailed study of the conditions that make blooms more likely to occur.
Fallon said the PKZILLA genes the team discovered are the first genes ever causally linked to the production of any marine toxin in the polyether group that prymnesin is part of.
Next, the researchers hope to apply the non-standard screening techniques they used to find the PKZILLA genes to other species that produce polyether toxins. If they can find the genes behind other polyether toxins, such as ciguatoxin which may affect up to 500,000 people annually, it would open up the same genetic monitoring possibilities for a suite of other toxic algal blooms with significant global impacts.
In addition to Fallon, Moore and Shende from Scripps, David Gonzalez and Igor Wierzbikci of UC San Diego along with Amanda Pendleton, Nathan Watervoort, Robert Auber and Jennifer Wisecaver of Purdue University co-authored the study.
Supporting documents
Click link to download and view these fileslow-res (18)
Other, FileSizeText 45.02 kb







No comments yet