The objectives of this study, published in Zoonoses, were to determine the prevalence of Cryptosporidium spp. infections in diarrheic calves reared in different localities in Egypt under different management systems, to clarify the role of the associated epidemiologic risk factors, and to identify the circulating Cryptosporidium spp. on molecular basis.
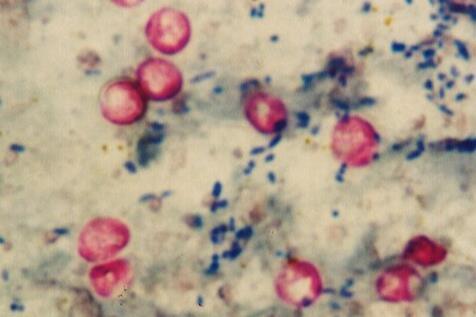
A total of 222 diarrheic calves (180 cattle and 42 buffaloes) were used in this study. The presence of the protozoan was proven by microscopic screening of the oocysts using a modified Ziehl-Neelsen staining technique followed by two-step nested PCR for the gp60 gene.
READ MORE: Gut bacteria protects against diarrhoeal disease
READ MORE: Scientists unveil cholangiocyte organoid system for Cryptosporidium parvum cultivation
Cryptosporidium oocysts were detected microscopically in 75 of 222 (33.78%) fecal samples, of which 71 (39.44 %) fecal samples were obtained from cattle calves and 4 (9.52 %) fecal samples were obtained from buffalo calves.
Uncommon subtype
The gp60 gene-based PCR was positive in 63 (84%) fecal samples, of which 59 (83.1%) fecal samples were obtained from cattle calves and 4 (100 %) fecal samples were obtained from buffalo calves. The sequences belonged to C. parvum subtype family IIa; an important zoonotic C. parvum.
C. parvum isolates in this study belonged to an uncommon C. parvum subtype family, especially ACATCA preceding the trinucleotide repeat. Different risk epidemiologic factors were verified to influence the prevalence of Cryptosporidium spp. infections.
The high prevalence recorded and the enzootic nature of cryptosporidiosis in calves in different localities in Egypt are associated with lack of hygienic measures as well as different epidemiologic variables. Further study is needed to illustrate the effect of heterogenicity of the obtained sequences on the biology of the parasite with public health significance.







No comments yet