A host of new antimicrobial strategies are in the development pipeline that could provide hope for healthcare sectors battling multi-drug resistant Staphylococcus aureus infections.
This wealth of candidates is explored in a new review, ‘Novel antimicrobial strategies to treat multi-drug resistant Staphylococcus aureus infection’, which has been accepted in Microbial Biotechnology, an Applied Microbiology International publication.
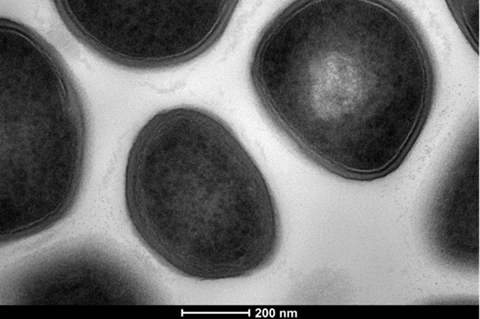
“There is a scarcity of novel anti-staphylococcal antibiotics hitting the clinic,” explains corresponding author Dr Edward Douglas from the University of Bath.
“In the last 20 years, only two truly novel antibiotic classes have been successfully licensed, the ribosomal targeting oxazolidinone linezolid, and the cell envelope targeting lipopeptide daptomycin. All additional antibiotics that have been successfully licensed for therapeutic use have been derivations of existing antibiotic classes such as β-lactams, tetracyclines, fluoroquinolones, glycopeptides and oxazolidinones.
“While modifications to these structures have been designed to retain activity against well-established resistance mechanisms, relying on this strategy may only delay the antibiotic resistance wave rather than overcome it.”
Tide is turning
However, it’s not all doom and gloom, and the tide is beginning to turn, he says.
“Recent government, pharmaceutical and charitable economic initiatives such as CARB-X, GARD-P as well as a $1 billion AMR action fund have been launched to try and reignite the antimicrobial development pipeline. Therefore, we were interested to see what novel antimicrobial strategies were currently being researched for use against S. aureus,” he says.
The team carried out a literature search to identify novel antimicrobial strategies in various stages of pre-clinical and clinical development, identifying companies that currently have an anti-infectives pipeline as well as searching the ClinicalTrials.gov registry using search terms related to S. aureus infection.
They found that significant biotechnological and scientific advances have been paving the way for the identification of the next generation of anti-staphylococcal antibiotics, epitomised by the renewed focus on developing new naturally occurring antibiotics.
“Newly developed analytic tools for effective genomic manipulation as well as our understanding of the structure of biosynthetic gene clusters (BCGs) has helped identify complestatin/corbomycin and malacidins,” Dr Douglas says.
“Searching for BCGs in previously unexplored locations such as the human microbiome, as well as the development of platforms to facilitate the isolation of previously unculturable bacterial species has yielded teixobactin, lugdunin and the humimycins.
“Similarly, improvements in bioengineering capabilities and our comprehension of how antimicrobial peptides (AMPs) exert their bactericidal activity is facilitating the design of synthetic AMPs such as PLG0206, OP-145, LTX-109 and AMP mimetics such as brilacidin.”
Selectivity towards bacterial cells
Dr Douglas says an important aspect of any prospective antibiotic is the selectivity towards bacterial cells over eukaryotic cells.
“Our understanding of S. aureus biology is ever improving and is helping to recognize essential proteins, not present in humans, that could be exploited by a therapeutic agent,” he says.
“For example, the LTA polymerisation enzyme LtaS, the enoyl-acyl carrier protein (ACP) reductase FabI, and the cell division apparatus protein FtsZ have all recently been identified as essential proteins that govern distinct bacterial processes. The ability of scientists to perform improved high throughput screens of chemical libraries and ligand based structural modelling has already proved fruitful in identifying inhibitors of these proteins.”
Such has been the success of β-lactamase inhibitors such as clavulanic acid, tazobactam, avibactam and so on in restoring susceptibility towards existing antibiotics that scientists are focused on recreating this and producing new antibiotic adjuvants against S. aureus.
“An MprF targeting antibody has been designed that has been shown to reverse MprF-mediated daptomycin resistance,” Dr Douglas says.
Drug repurposing programmes
“Drug repurposing programmes have also identified the tyrosine kinase inhibitor nilotinib as a potent NorA inhibitor capable of restoring ciprofloxacin susceptibility.
“β-lactams such as methicillin remain an antibiotic mainstay in the treatment of S. aureus - however resistance to this is achieved through the acquisition of an alternative penicillin binding protein (PBP2A) which gives rise to notorious MRSA strains.
“Reversal of this type of this resistance is a significant area of research. Strategies to achieve this include the mecA antisense oligonucleotide PS-ODN04, which eradicates the expression of PBP2A. Numerous other molecules that potentiate the activity of β-lactams have been identified and include polyamines, statins, SpsB inhibitors and nucleoside analogues.”
Due to the threat of AMR and the impressive adaptability of S. aureus, suitable contingencies need to be in place that don’t rely on the use of conventional antibiotics to treat resistant infections.
Promise of phages
“Bacteriophages and the lytic enzymes or ‘enzybiotics’ they produce to kill their bacterial hosts are a very promising contingency strategy,” Dr Douglas says.
“A significant advantage of bacteriophages is their precision and narrow spectrum of activity which can substantially limit off target damage to the microbiome.
“The compassionate use of bacteriophages has largely been associated with positive clinical outcomes, which has provided much of the rationale leading to the initiation of a number of bacteriophage clinical trials. Likewise, the tonabacase and exebacase phage lysins have shown promise in early clinical trials.”
This study was led by Dr Edward Douglas and Dr Maisem Laabei and supported by Sri Wijayanti Wulandari and Dr Scott Lovell, all of the Department of Life sciences at the University of Bath.
‘Novel antimicrobial strategies to treat multi-drug resistant Staphylococcus aureus infection’, appears in Microbial Biotechnology.
Supporting documents
Click link to download and view these filesG3_sample4_019
Image, FileSizeText 16.5 mbA3_sample1_049
Image, FileSizeText 16.5 mbA3_sample1_LM
Image, FileSizeText 16.5 mbA3_sample1_038
Image, FileSizeText 16.5 mb






No comments yet