Researchers have developed a fast and highly efficient engineered pathogen capture method which shortcuts the accurate detection of pathogens in small blood samples, potentially saving lives.
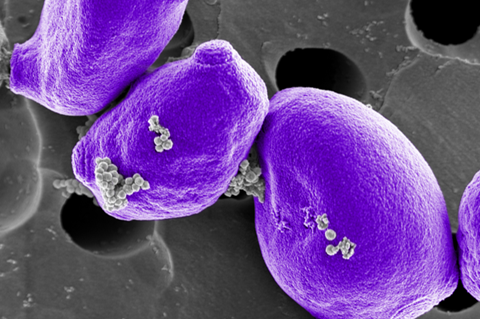
The collaborative team led by researchers from Great Ormond Street Institute of Child Health (GOSH), London and including researchers from the Wyss Institute for Biologically Inspired Engineering at Harvard University and BOA Biomedical in Cambridge has re-engineered the process of microbial pathogen identification in blood samples from pediatric sepsis patients using the Wyss Institute’s FcMBL broad-spectrum pathogen capture technology.
The advance enables accurate pathogen detection with a combination of unprecedented sensitivity and speed, and could significantly improve clinical outcomes for pediatric and older patients with bloodstream infections (BSIs) and sepsis. The findings were published in PLoS ONE as ’FcMBL magnetic bead-based MALDI-TOF MS rapidly identifies paediatric blood stream infections from positive blood cultures’.
BSIs with various microbial pathogens can rapidly escalate to life-threatening sepsis when the body is overwhelmed by the multiplying invaders and shuts down its organs’ functions. In 2017, there were 48.9 million cases and 11 million sepsis-related deaths worldwide. Almost half of all global sepsis cases occurred among children, with an estimated 20 million cases and 2.9 million global deaths in those under five years of age.
Need for speed
To prevent BSIs from progressing to full-blown sepsis, the infection-causing bacterial or fungal species must be identified as fast as possible. Only then can optimal pathogen-tailored antibacterial or antifungal treatments be applied in time. The conventional method used in clinical laboratories to identify the causative pathogenic species is long and laborious, requiring two time-consuming culture steps that take at least 1 to 3 days to complete.
“For all patients with sepsis, their chances of surviving dramatically shrink the longer it takes to identify the infection-causing pathogen(s) and thus, receive the most promising antimicrobial treatment,” said Nigel Klein, a Professor of Infectious Disease and Immunology at GOSH, and a senior author on the study.
“At Great Ormond Street Hospital we have been working to demonstrate both the importance rapid diagnosis and the fact that with innovative approaches we can identify the causative organism in between 40 minutes and six hours. Compared to adult patients, sepsis in infants and small children progresses much faster, and therefore there is a real need for diagnostic methods that support early detection. Accurate diagnosis is even more significant due to the availability of only small blood volumes from paediatric patients which can make re-sampling challenging.”
In 2020, senior authors Klein and Elaine Cloutman-Green, PhD, a Consultant Clinical Scientist and Infection Control Doctor at GOSH, began collaborating with Lead Staff Scientist Michael Super, Ph.D. and Founding Director Donald Ingber, M.D., Ph.D. at Harvard’s Wyss Institute to solve this problem.
“Based on our earlier success with FcMBL in isolating pathogens from joints as well as bovine and human blood with extraordinary efficiencies, we hypothesized that building an FcMBL-mediated pathogen capture into a modified clinical blood culture protocol could shorten the time and reduce the size of the required patient samples to yield the same results that time-consuming blood culture protocols provide,” said Super.
Current methods
In the pathogen identification process currently performed in clinical settings, blood samples are first added to bottles containing liquid media in which infectious microbes, if present, are amplified to a certain density. Then, the amplified microbes are grown on solid media as isolated colonies whose constituent cells eventually can be identified with a highly sensitive, yet fast and relatively inexpensive analytical method know as MALDI-TOF mass spectrometry (MS).
“Indeed, isolating the infectious microbes directly from grown liquid blood cultures using FcMBL makes them available for MALDI-TOF MS analysis much earlier,” added Super.
FcMBL is the key component of a broad-spectrum pathogen capture technology. It consists of a genetically engineered human immune protein called mannose-binding lectin (MBL) that is fused to the Fc fragment of an antibody molecule to produce the resulting FcMBL protein.
In this configuration, the MBL portion of FcMBL can capture more than 100 different microbial species with high efficiency, including virtually all of the bacterial and fungal pathogens causing sepsis. FcMBL’s Fc portion can be used to couple it to magnetic beads, allowing the captured pathogens to be quickly pulled out of patient samples and liquid blood cultures.
Early stages
In the earlier stages of the project, the Wyss team provided purified bead-coupled FcMBL to the GOSH team, which had access to blood samples from paediatric patients at the hospital. At later stages, the sepsis and infectious disease company BOA Biomedical, co-founded by Super and Ingber to commercialize the Wyss Institute’s FcMBL technology, provided the FcMBL reagent and critical expertise to the project. BOA Biomedical meanwhile developed the manufacturing capabilities for FcMBL that the Food and Drug Administration (FDA) in the US and other federal health agencies require for producing therapeutic and diagnostic products.
“Sepsis is the leading killer in hospitals, and rapidly initiating the right antibiotic saves lives. Using work originally developed at the Wyss Institute, BOA Biomedical’s revolutionary FcMBL technology helps to quickly and accurately identify the pathogen causing sepsis, ushering in a new era of targeted antimicrobial therapy to help individual patients and curb society’s deadly antimicrobial resistance problem,” said Mike McCurdy, Chief Medical Officer of BOA Biomedical.
In addition to using the gold standard two-step blood culture in combination with MALDI-TOF MS pathogen identification, the team also included the Bruker Corporation’s MBT Sepsityper® kit as a comparison. Brought to market in 2021, the MBT Sepsityper® essentially eliminates the time-consuming second microbial culture step by lysing microbial cells from the liquid culture and spinning the fragments down in a centrifuge before analyzing them by MALDI-TOF mass spectrometry analysis.
Although it accelerates the overall diagnostic process, the MBT Sepsityper® method produces lower microbial detection rates than those obtained with the conventional culture method, which means that it may still fail to identify the infection-causing pathogen in a significant fraction of blood samples.
New opportunities
“Our FcMBL approach has opened up the opportunity to identify pathogenic organisms to guide treatment 24 to 48 hours earlier than would be possible using standard culture techniques. It has also enabled us to use this identification to make any ongoing culture for antibiotic sensitivities more tailored to the needs of the patient. This method isn’t tied into a specific platform or manufacturer, and thus we see clear potential for it to become a new standard processing step for clinical pathogen detection,” said Cloutman-Green.
“The FcMBL method identified 94.1% of microbial species found in clinical blood culture analysis with samples from 68 pediatric patients,” said first author Kerry Kite, who performed her graduate work with Klein and Cloutman-Green.
“We were able to identify more infectious species in positive liquid blood cultures using the FcMBL method than with the MBT Sepsityper® method (25 of 25 vs 17 of 25), and this trend was even further pronounced in the case of the common fungal pathogen Candida (24 of 24 vs 9 of 24).”
Candida species account for about 5% of all cases of severe sepsis and are the fourth most common pathogen isolated from patients’ bloodstreams in the United States. Not only do infections with Candida and other fungi require specific antifungal treatments, distinguishing among the various types of pathogenic fungi helps direct the appropriate antimicrobial therapy. Specifically in neonatal intensive care units, Candida infections are a major cause of morbidity and mortality, killing as many as 40% of infants and often causing neurodevelopmental impairments in those that survive.
“By continuously adapting the powerful FcMBL pathogen capture technology to unmet and pressing diagnostic needs, such as the rapid diagnosis of sepsis in pediatric patients, we hope to profoundly alter the frequently dismal prospects of patients of all ages,” said Ingber.
“Our ultimate goal is to be able to accurately and even more rapidly identify pathogens directly in small samples of blood without the need for any additional microbial cultures.”
Topics
- BOA Biomedical
- BSIs
- Cambridge
- Candida
- Donald Ingber
- Elaine Cloutman-Green
- Fungal Therapeutics
- Fungi
- Great Ormond Street Hospital
- Great Ormond Street Institute of Child Health
- Harvard
- Infection Prevention & Control
- Infectious Disease
- Innovation News
- Kerry Kite
- MBT Sepsityper
- Michael Super
- Nigel Klein
- One Health
- pathogen identification
- PLOS One
- Public Health
- sepsis
- UK & Rest of Europe
- USA & Canada
- Wyss Institute for Biologically Inspired Engineering




No comments yet