For millions of years, underground fungi have lived in symbiosis with plant roots. Plants provide photosynthesized carbon, while fungi deliver water and nutrients. In order to do so, these organisms share space at cellular scale: fungi stretch a network of tendrils called arbuscules into a plant’s root cells, and both organisms rearrange their cells around this structure to facilitate sharing.
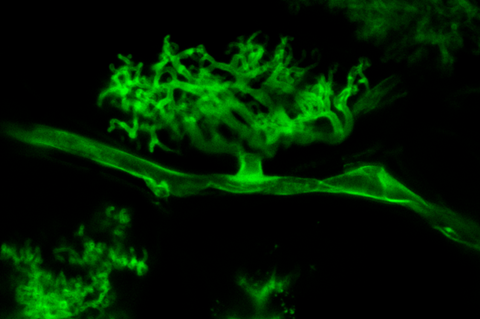
Recently, researchers have been able to study both sides of this interaction up close, using RNA sequencing to understand gene expression: one of the first cross-kingdom spatially-resolved transcriptomics studies to date. This paper is the cover article for the April 2024 issue of Nature Plants.
“We wanted to better understand the nature of this symbiosis at the cellular level — really understand how those two cell types [of two different organisms] are interacting with each other, without all the noise or other biological activity of the surrounding neighborhood,” said Benjamin Cole, senior author of this work. Cole is a research scientist at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab).
Reservoir for carbon compounds
Specific understanding of this symbiosis could offer improvements in multiple directions. On the fungi side, underground mycorrhizal networks can serve as a reservoir for carbon compounds that plants generate from carbon dioxide they take in. In this way, encouraging this symbiosis could improve the way soils store carbon from the atmosphere. For plants, boosting this relationship could improve biofuel feedstocks growing in nutrient-poor fields.
“This is one interaction that allows plants to better survive in those environments,” Karen Serrano said. She is first author of this paper and a graduate researcher at the Joint BioEnergy Institute (JBEI).
Cole led the work in collaboration with JBEI as part of the 2021 Laboratory Directed Research and Development (LDRD) program. Cole also received a DOE Early Career Research Program (ECRP) award in 2021, which aims in part to build off of this work. Former JGI postdoctoral scholar Margot Bezrutczyk collaborated extensively with Cole and Serrano to collect and analyze the single nuclei and spatial transcriptomics data this work leverages.
Two for one
These experiments focused on two species at once: the model legume species Medicago truncatula and the mycorrhizal fungi Rhizophagus irregularis. To see how these organisms cooperate, this team applied R. irregularis spores directly to M. truncatula seedlings grown in a controlled environment chamber, so the fungus could colonize the plants’ roots. Then, comparing with control seedlings which were not treated with fungi, they used multiple approaches to look at gene expression in both plant cells and fungal cells.
Using a technique called single nuclei RNA sequencing, the researchers identified different cell types within M. truncatula root cells, and profiled their gene expression. Then, the researchers used a technique called spatial transcriptomics, to generate maps of gene expression. This spatial transcriptomics technique allowed them to understand gene expression within circular capture areas roughly 55-microns in diameter — about the width of a human hair. At such resolution, this spatial transcriptomics data captured molecular information from both plant cells and fungal cells.
“Because this technology relies on just polyadenylated transcript capture — any RNA that is from eukaryotes — we were able to capture both plant and fungal transcripts,” Serrano said. This team quantified the expression of over 12,000 fungal genes, in addition to associated plant genes.
Tuning the symbiosis
All together, these data offer a granular view of both plant and fungal activity at different stages of this symbiosis. Within that activity, Serrano, Cole and their team found over 1,000 upregulated genes, 188 of which were shared with previous studies in the same system. With the right functional characterization, those genes could become dials for tuning this symbiosis. “Those are great candidates for genetic engineering. Our hope is that the community at large will follow that up,” Serrano said.
This work focused on a relatively well-understood model system, so future directions will also include targeting biofuel feedstocks in similar studies. “We would like to look at arbuscular mycorrhizal symbiosis in other bioenergy grasses, like sorghum and switchgrass. We’re optimizing systems now, so that we can get those working,” Cole said.




No comments yet