A new study identifies a gene in a rough Salmonella Infantis variant that could offer a target for developing effective vaccines and therapeutic strategies.
And because the gene has a high homology with other serotypes and those of other members of the Enterobacteriaceae family, it could provide a broad therapeutic intervention for other pathogens as well, the authors suggest.
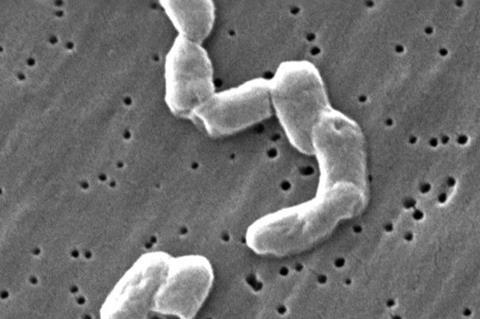
Led by AMI member Nneka Vivian Iduu, the study, ‘Mutation in Wzz(fepE) Linked to Altered O-Antigen Biosynthesis and Attenuated Virulence in Rough Salmonella Infantis Variant’ was published in Veterinary Sciences on November 28 2024.
“The primary goal of our study was to characterize the genomic features of a rough Salmonella Infantis variant and determine the genetic changes that affect its virulence. We identified that the loss of function of the Wzz(fepE) gene, which regulates the length of the O-antigen, is likely linked to altered O-antigen biosynthesis. This alteration appears to contribute to the reduced virulence observed in the rough strain,” Nneka said.
Significant foodborne pathogen
Salmonella Infantis has emerged as a significant foodborne pathogen worldwide, particularly in poultry. Its prevalence is rising, driven by antimicrobial resistance and its zoonotic potential, making it a pressing global health concern.
READ MORE: Rising antimicrobial resistance in some Salmonella serovars isolated from retail chicken meat
READ MORE: Flour power: why it’s not a great idea to eat it raw
The ability of this pathogen to evade host immune defenses and establish infections is closely tied to its virulence factors, including those involved in O-antigen biosynthesis. The identification and characterization of genetic contributors to its pathogenicity are critical for devising targeted interventions to mitigate its impact. These efforts hold promise for the development of glycoconjugate vaccines, which could leverage structural modifications in the O-antigen to enhance immune protection and improve food safety measures.
Genomics techniques
Nneka employed state-of-the-art whole genome sequencing and comparative genomics techniques to examine the genetic makeup of smooth and rough S. infantis strains. Recognizing the integral role of O-antigen biosynthesis genes in virulence, the team focused their analysis on these genes.
“While the smooth and rough strains displayed high levels of genomic similarity, we identified a frameshift mutation in the Wzz(fepE) gene of the rough variant. This mutation resulted in premature truncation of the protein, which was hypothesized to be the primary cause of the altered O-antigen biosynthesis,” she said.
“What was more surprising was that both strains lost similar O-antigen biosynthesis genes but this did not seem to affect the virulence of the smooth strain when a chicken embryo lethality assay was conducted to evaluate the pathogenicity of both strains. In contrast, the rough variant showed a significantly lower virulence in similar experimental conditions.
Foundation for vaccines
“These findings highlight the importance of the Wzz(fepE) gene in S. Infantis virulence and suggest that targeting this gene could provide a foundation for developing effective vaccines and therapeutic strategies. In addition, being that the gene has a high homology with other serotypes and those of other members of the Enterobacteriaceae family, it could provide a broad therapeutic intervention.”
To build on these findings, Nneka intends to further investigate the specific role of Wzz(fepE) in Salmonella virulence by using CRISPR/Cas9 gene editing to conduct reverse genetics. This involves altering the gene in the smooth strain of S. Infantis as well as in other Salmonella serotypes to assess phenotypic changes and validate these findings across different host environments.
“Upon successful confirmation, I plan to evaluate the protective efficacy of the attenuated strains by challenging them with virulent strains in animal models. I believe that developing experimental vaccines targeting this pathway represents a promising direction for future research,” Nneka said.
This study was led by Nneka Vivian Iduu under the supervision of Dr. Chengming Wang from Auburn University, Auburn Alabama, USA. The work was funded by the USDA Agricultural Research Service Program.






No comments yet