Pioneering research utilizes high-throughput single-cell sequencing to demystify the microbial universe within activated sludge, a cornerstone of wastewater treatment. This study has unearthed a plethora of antibiotic resistance genes (ARGs) and has pointed to the existence of previously unknown microbial species, significantly advancing our comprehension of microbial diversity and the mechanisms of genetic exchange within this environment.

READ MORE: Wastewater monitoring can detcet foodborne illness
READ MORE: New antibiotic resistant species of bacteria found in hospital wastewater system
Activated sludge is essential for removing contaminants from wastewater, and understanding the diverse microbial communities it harbors has been a significant task. Traditional methods fail to capture the full spectrum of microbial interactions, especially the minority.
Due to these limitations, researchers recognized the need for single-cell level investigations to gain deeper insights into microbial heterogeneity.
Intricate genetic networks
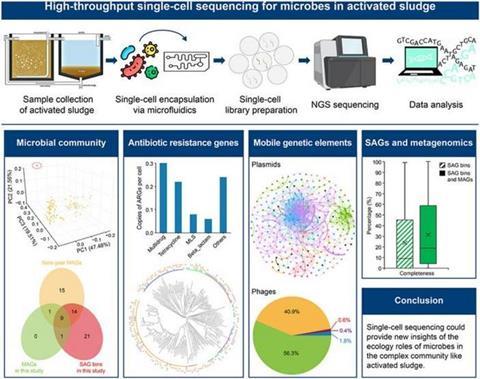
Led by the University of Hong Kong and published in Environmental Science and Ecotechnology, the study applied single-cell sequencing to samples from a Hong Kong wastewater plant. This method overcame the constraints of traditional metagenomics, revealing the intricate genetic networks within microbial communities and offering new perspectives on antibiotic resistance gene transfer.
The research team sequenced 15,110 individual microbial cells, classifying them into 2,454 single-amplified genome bins and discovering that 27.5% represent previously unclassified species.
The study shed light on microbial dark matter and offered new insights into the complexity of microbial ecosystems. The identification of 1,137 ARGs, along with numerous plasmids and phages, underscored the frequent horizontal gene transfer between species.
Plasmids were found to be instrumental in spreading ARGs, raising concerns over the potential spread of antibiotic resistance. The findings highlight the need for closer monitoring of wastewater systems to prevent public health risks.
Highlights
•This study marks the first application of high-throughput single-cell sequencing to analyze the activated sludge (AS) microbiome.
•A total of 15,110 single cells were sequenced, resulting in 2,454 single-amplified genome bins.
•The analysis detected 1,137 antibiotic resistance genes, 10,450 plasmid fragments, and 1,343 phage contigs.
•The shared relationships for plasmid (12,819) and phage (184) hosts indicate a high frequency of horizontal gene transfer.
•A set of 1,529 metagenome-assembled genomes was used to further classify 98 previously unclassified SAG bins.
Prof. Tong Zhang, the lead researcher, emphasized the study’s importance: “Our research highlights the power of single-cell sequencing in revealing the hidden dynamics within wastewater treatment. These findings on antibiotic resistance gene transfer pave the way for improved monitoring and mitigation strategies, which are crucial for safeguarding public health under a One Health approach.”
The research has wide-reaching implications, particularly in enhancing wastewater treatment processes and curbing the spread of antibiotic resistance. The ability to track ARGs at a single-cell level could influence future environmental policies and public health measures, while also opening new avenues for applications in other ecosystems, such as soil and the human gut.







No comments yet