Researchers at Nano Life Science Institute (WPI-NanoLSI), Kanazawa University, IMDEA Nanoscience (Madrid, Spain) and CNB-CSIC (Madrid, Spain) report in ACS Nano experiments that reveal a cycle of conformational stages that recombinant Influenza A genomes pass through during RNA synthesis.
Influenza A is a global health risk responsible for local epidemics and deadly pandemics. As such, the mechanism by which the virus replicates itself has attracted significant interest.
READ MORE: Study shows promise for a universal influenza vaccine
READ MORE: Influenza viruses can use two ways to infect cells, study finds
Researchers led by Shingo Fukuda at Nano Life Science Institute (WPI-NanoLSI) at Kanazawa University in Japan, Jaime Martin-Benito at CNB-CSIC and Borja Ibarra at IMDEA Nanociencia in Spain, used high-speed atomic force microscopy and electron microscopy to pin down the conformational dynamics of recombinant viral genomes (or rRNPs) during RNA synthesis.
Bulky double-helical structure
Previous attempts to understand what possible conformational changes occur during Influenza A viral multiplication cycle had been hindered by what the researchers describe as the “bulky double-helical structure” of the viral RNPs (vRNP, Fig.1 A), which made it hard to see what was going on. As a result the researchers produced a circular recombinant ribonucleoprotein complex (rRNP) (Fig.1 B), which allowed them to overcome the ‘bulky issue’. The authors used HS-AFM to follow conformational changes of individual rRNP complexes in real-time during active RNA synthesis (Fig.1 C).
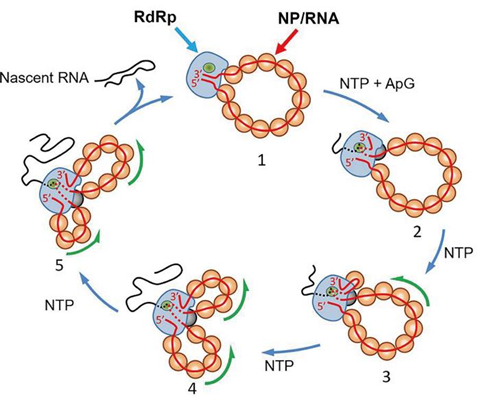
Their work provides first direct experimental evidence showing that individual rRNPs can be recycled for multiple transcription and replication cycles. This is a key feature for viral multiplication. In addition, their study highlights how factors that affect the stability of the secondary structures of the nascent RNA affect the rate of RNA synthesis.
Valuable approach
The authors concluded that the approach is useful for investigating viral transcription and replication mechanisms. “Transcriptional pausing is an intrinsic property of most RNA polymerases, and its regulation constitutes one of the central mechanisms of control of gene expression,” they add.
”Future single-molecule experiments of real-time RNA synthesis kinetics by the IAV RdRp [influenza A virus RNA dependent RNA polymerase] within the context of the RNP will help to elucidate the nature of putative pause states and their roles in viral transcription and replication.”
Background
Ribonucleic acid (RNA) is a polymer and a nucleic acid. Its replication is catalyzed by an enzyme known as RNA polymerase. The synthesis only proceeds in the presence of the nucleotide and proteins required to build up the RNA molecule.
Although RNA synthesis can use DNA as the synthesis template, a number of viruses replicate with RNA as the template. The enzyme that catalyzes this type of synthesis is known as RNA dependent RNA polymerase.
Recombinant ribonucleoproteins are useful for ways of studying RNA processes. In this instance the authors used a recombinant ribonucleoprotein made up of the same protein components but just 352 nucleotides long, where the RNA segment used had been shown to avoid supercoiling.
High-speed atomic force microscopy
High-speed atomic force microscopy uses a nanosized tip at the end of a cantilever that is scanned over a sample. It can be used to determine the topography of a sample surface from the change in the strength of forces between the tip and the sample with distance, and the resulting deflection of the cantilever. It was first developed in the 1980s but a number of modifications have augmented the functionality of the technique since. It is better suited to imaging biological samples than the scanning tunneling microscope that had been developed because it does not require a conducting sample.
In the 2000s Toshio Ando at Kanazawa University was able to improve the scanning speed to such an extent that moving images could be captured. This allowed people to use the technique to visualize molecular processes for the first time.






No comments yet