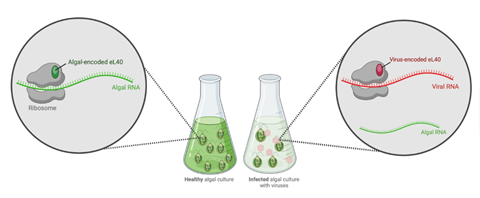
Researchers at the University of Hawai‘i at Mānoa recently discovered that a virus, FloV-SA2, encodes one of the proteins needed to make ribosomes, the central engines in all cells that translate genetic information into proteins, the building blocks of life. This is the first eukaryotic virus (a virus that infects eukaryotes, such as plants, animals, fungi) found to encode such a protein.

Viruses are packets of genetic material surrounded by a protein coating. They replicate by getting inside of a cell where they take over the cell’s replication machinery and direct it to make more viruses. Simple viruses depend almost exclusively on material and machinery provided by the host cell, but larger, more complex viruses code for numerous proteins to aid in their own replication.
“We were excited to discover that this virus encodes a ribosomal protein called eL40,” said Julie Thomy, lead author of the study and postdoctoral researcher in the Daniel K. Inouye Center for Microbial Oceanography: Research and Education (C-MORE) and Department of Oceanography in the UH Mānoa School of Ocean and Earth Science and Technology (SOEST). “It makes sense that a virus could benefit from altering this critical piece of cell machinery, but there was just no evidence for it in any eukaryotic virus.”
Isolating new viruses
The virus was discovered as part of a larger effort by members of the Marine Viral Ecology Laboratories (MarVEL) in SOEST to isolate and characterize new viruses that live in the ocean. A former Oceanography graduate student, Christopher Schvarcz, sampled water from Station ALOHA 60 miles north of O’ahu, Hawai‘i, and subsequently isolated dozens of viruses. Among them was FloV-SA2, which infects a species of phytoplankton called Florenciella.
“Chris was so productive at isolating viruses, he could not analyze them all before he left,” said Grieg Steward, Oceanography faculty member overseeing the project. “Detailed analysis of this virus had to wait until Dr. Thomy joined the lab, but it was worth the wait!”
Preferential production
Previous discoveries have shown that, like FloV-SA2, other so-called ‘giant’ viruses code for proteins involved in a wide range of metabolic processes. Some, such as those involved in fermentation or sensing light, seem like surprising functions to find in a virus. These genes must help the virus replicate, but, as is the case with the ribosomal protein, it is not always clear how. The researchers are now focused on figuring out the details of how and when this protein is used by the virus.
“Our working hypothesis is that by inserting one of its own proteins into the ribosome, the virus alters this key piece of machinery to favor the production of virus proteins, over the usual cell proteins,” said Thomy.
Ocean ecosystems
“Viruses are integral to the functioning of ocean ecosystems, influencing biological productivity, shifting community interactions, and driving evolutionary change,” said Steward. “This discovery reveals new details about the complex ways viruses in the ocean interact with phytoplankton, which are the foundation of ocean ecosystems, but it also opens new avenues in our understanding of the fundamentals of viral biology.”
The scientists expect that FloV-SA2 will be a valuable model system for investigating new mechanisms by which viruses manipulate cell metabolism and redirect host resources and energy.






No comments yet