A new group of mitochondrial viruses confined to the arbuscular mycorrhizal fungi Glomeromycotina may represent an ancestral lineage of mitoviruses.
Bacteriophages are viruses that infect bacteria. As former bacteria, there are also viruses that infect mitochondria, known as mitoviruses, which evolved from bacteriophages. While mitoviruses have been found in fungi, plants, and invertebrates, they are not well studied.
Associate Professor Tatsuhiro Ezawa at Hokkaido University, Professor Luisa Lanfranco at University of Turin, and Dr. Massimo Turina at National Research Council of Italy (CNR) Torino led an international team to discover a new group of mitoviruses, called large duamitoviruses. Their findings were published in the journal mBio.
“In their current form, mitoviruses are RNA molecules within mitochondria that encode only the RNA-dependent RNA polymerase (RdRp) used for genome replication,” explains Ezawa. “They are hypothesized to affect the virulence of plant pathogens and plant resilience to abiotic stress. Most interestingly, mitoviruses are transmitted not only vertically to progeny via mitochondrial division but occasionally also horizontally betwneen distant species.”
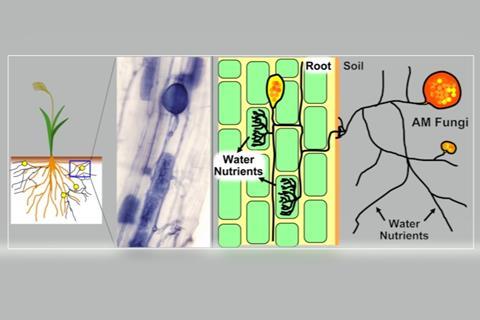
The team analyzed the RdRp enzyme from 10 new mitoviruses and sequences from previous research and public databases. This analysis revealed the existence of peculiar large duamitoviruses that are exclusive to the Glomeromycotina, a group of mycorrhizal fungi which are very widespread in nature and provide several benefits to the host plants.
Unusual characteristics
These large duamitoviruses possess two structurally distinct characteristics: they encode larger than average RdRp (~1,036 amino acids long) with a unique amino acid motif, and the UGA codon is rarer than in other mitoviruses. Furthermore, a phylogenetic analysis showed that the large duamitoviruses were evolutionarily distinct from other mitoviruses and likely represent an ancestral lineage.
“One of our most interesting discoveries is that the large duamitoviruses appear to be exclusive to glomeromycotina,” Lanfranco described. “We analysed the global distribution of all the mitovirus RdRp sequences included in our study, and we found that large duamitoviruses were globally distributed in ecological niches occupied by glomeromycotinian fungi. Although other fungi are found in these niches, all currently available large duamitoviral sequences could be only associated with glomeromycotinian fungi.”
The team hypothesizes that there is a transmission barrier that prevents the horizontal transfer of large duamitoviruses. Future work will focus on understanding these barriers, on confirming that large duamitoviruses represent an ancestral lineage of mitoviruses, as well as elucidating the functional significance of their exclusive presence in glomeromycotina.




No comments yet