Thanks to support from Applied Microbiology International, scientists testing sampling collection protocols in Mars analogue conditions have shown that non-scientists will be able to replicate the tests as long as they follow the methods.
With the help of AMI’s Small Research Projects Equipment Fund, the team at the Mars Desert Research Station were able to show that portable lab equipment could be used in Martian analogues to investigate the presence and functions of microorganisms in extreme environments.

The grant was awarded to Daniel Loy of The Open University for his project ’Investigating the use of portable laboratory equipment at the Mars Desert Research Station’ as part of his student placement for his PhD, with collaboration from all of MDRS Crew 292. His PhD supervisor is Dr Michael Macey of The Open University.
Mars research
He explained: ”This project was carried out at the Mars Desert Research Station (MDRS), a Martian analogue research station run by The Mars Society in Utah, USA.
”MDRS Crew 292 Mangalyatri (Mars Explorers) was a primarily Indian national crew brought together by Mars Society Australia with the aim of bringing the experience and lessons learned from the MDRS back to India to inform the building of a new Science Desert Research Station in the high glacial deserts of the Himalayas in the Ladakh region of India.

”My objective as the Crew Biologist during this mission was to conduct scientific operation research on the use of portable equipment in an analogue environment: carrying out DNA extractions, PCR and gel visualizations with the Bento Lab portable PCR workstation. This is a combined portable centrifuge, PCR machine and gel imaging station that was used in conjunction with a Qiagen DNeasy Powersoil Pro DNA extraction kit.”
Simulation conditions
The mission was conducted in simulation conditions, so in order to leave the crew habitat and collect samples, the team members had to wear Extra Vehicular Activity (EVA) suits.
”These made the collection of samples more difficult but with the use of sterilised digging tools, sterile cotton wool buds, sample bags and falcon tubes samples were collected by multiple crew members across different EVAs,” Daniel said.

As the purpose of this project was to test the operational capability, rather than find novel species or communities, samples were collected from the most likely places to contain active microorganisms seen on EVAs. Samples were collected from soil in sheltered conditions such as between large rocks and under rocks where wind had eroded the surrounding softer topsoil, as well as opportunistically sampling any small bodies of water (Figure 1).
DNA extraction
These samples were brought back to the Science Dome where DNA extractions were performed in a laminar flow cabinet, using the Powersoil Pro kit and the centrifuge in the Bento Lab.

”The remote nature of this project meant that it was vital to be adaptable - as there was no vortex or other shaking equipment available to agitate samples and lyse cells as the first part of the DNA extraction protocol, they had to be shaken by hand,” Daniel said.
”The protocol called for 10 minutes of horizontal shaking so samples were shaken by hand for 12 minutes due to the difference in speeds. This may have been the limiting factor influencing the success of this project, as only two soil samples had DNA successfully extracted and quantified using untargeted PCR.”
Presence of DNA
The untargeted PCR was used as an initial indicator of the presence of DNA through the observation of bands of amplified DNA at the top of the gel. The two samples that had DNA successfully extracted both came from soil that was collected from underneath rocks as highlighted below in Table 1.
Table 1. List of Samples/Cultures that had DNA extraction performed on them.
| Name | Type | Sol, EVA and Location |
|---|---|---|
|
L |
Lichen |
Sol 4, EVA 4 N4253168, E518918 |
|
G |
Green Mudstone |
Sol 4, EVA 4 N425376, E518972 |
|
S |
Soil under rock |
Sol 4, EVA 4 N4253311, E518982 |
|
WA |
Water |
Sol 6, EVA 7 N4251004, E518556 |
|
WB |
Water |
Sol 6, EVA 7 N4251131, E518493 |
|
S1 |
Soil |
Sol 7, EVA 8 N4252848, E518615 |
|
S2 |
Soil |
Sol 7, EVA 8 N4252922, E518410 |
|
C |
Soil from eroded crevice of rock |
Sol 8, EVA 9 N4249486, E518361 |
|
Y |
“Yellow goo” found in small body of water |
Sol 8, EVA 9 N4249488, E518287 |
|
Y2 |
“Yellow goo” found in small body of water, repeat extraction of above sample |
As above |
|
YSP |
“Yellow goo” inoculated in 1:10 dilution of SP media |
As above |
|
SSP |
Soil under rock inoculated in 1:10 dilution of SP media |
Sol 4, EVA 4 N4253311, E518982 |
|
SRN |
Soil under rock inoculated in RN media |
As above |
DNA extractions were carried out from 0.25 grams from each sample, or in the case of liquid samples/cultures 200ul of liquid after being agitated by hand for 10 seconds. The Powersoil Pro Kit Extraction protocol was followed for all extractions, with hand mixing being used to lyse and mix during the process, as no vortex was available. DNA was successfully extracted from two soil samples collected on different EVAs from different sites (table 1).
Using targeted PCR, archaea and fungi were identified to be within the soil in both S and S2 samples. The S soil sample also contained bacteria, while this could not be identified within the S2 sample due to an amplification failure.
Functional genes
The presence of functional genes was also investigated, with the large subunit gene hhyL of the group 1h [NiFe]-hydrogenases (imaged below) and the sulfide:quinone oxidoreductase gene fragment both found within DNA extracted from soils across the MDRS sampling sites. This provides an indication as to the functional potential of the resident microbiome and could be used to inform targeted enrichments for specific metabolisms in the future.
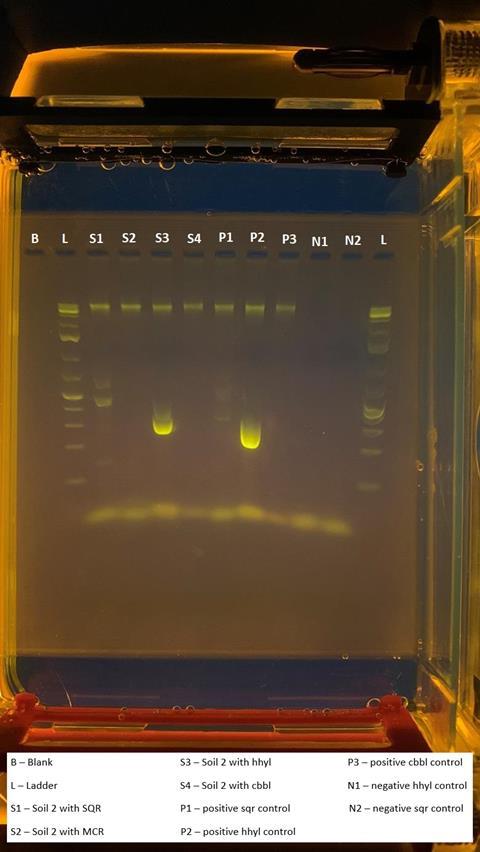
”The successful extraction, amplification and visualisation of DNA and specific genes with this equipment shows that portable laboratory equipment can be used in Martian analogues to investigate the presence and functions of microorganisms in other extreme environments,” Daniel said.
”Pre-existing protocols for all processes were followed, meaning anyone, including non-biologists, with the right equipment would be able to replicate these methods including at the proposed Ladakh analogue research station.
”This project would not have been possible without the funding provided from Applied Microbiology International through their Small Research Project and Equipment Grant. I would also like to thank the Central England NERC Training Alliance for funding my PhD and also assisting with the costs as this was my student placement,” Daniel said.
Topics
- Applied Microbiology International
- Archaea
- Asia & Oceania
- Bacteria
- Be inspired
- Community
- Daniel Loy
- Early Career Research
- Extremophiles
- Fungi
- Mars Desert Research Station
- MDRS Crew 292 Mangalyatri
- Michael Macey
- Microbes and Space
- Microbes and Space
- Microbes and Space
- Microbiological Methods
- Small Research Projects Equipment Fund
- space
- The Mars Society
- The Open University
- UK & Rest of Europe
- USA & Canada








No comments yet