A team of scientists has mapped the times during a batch culture when core biosynthetic genes surged into action, showing that bursts of biosynthetic gene cluster (BGC) transcriptional activity correlated with surges in net production rates per cell of known natural compounds.
The research, ‘Bursts in biosynthetic gene cluster transcription are accompanied by surges of natural compound production in the myxobacterium Sorangium sp.’ by a team from Leibniz Institute DSMZ appears in Microbial Biotechnology, an Applied Microbiology International publication.

They found that the core biosynthetic genes from most BGCs in the wild-type myxobacterium Sorangium sp. So ce836 were actively transcribed at specific time points in a batch culture. A majority of polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS) genes reached their maximal transcription in the exponential phase.
Strikingly, the bursts in BGC transcriptional activity correlated with surges in net production rates per cell of known natural compounds.
Mining for molecules
While natural compounds are a rich source of biologically active molecules for the discovery of novel drugs, such as antibiotics, and antifungal and antitumour drugs, the detection, identification, and production of such natural compounds in sufficient yield poses a difficult challenge, said corresponding author Prof. Dr. Ulrich Nübel, Head of the Microbial Genome Research unit at Leibniz Institute DSMZ.
“The number of BGCs in bacterial genomes commonly far exceeds the number of natural products detected in cultivated bacterial strains, suggesting enormous potential for novel compounds yet to be discovered,” he said.
Hidden potential
“Accordingly, it is frequently assumed that most BGCs may be ‘silent’ (not expressed), and that production of natural compounds occurs under specific conditions only, such as nutrient-limitation or other stress factors.
“However, the regulation of compound biosynthesis is little understood. Our study focused on the elucidation of the temporal transcriptional regulation and production of natural compounds in the wild-type myxobacterium Sorangium sp. So ce836.”
The researchers sequenced the genome of So ce836, annotated it, and by genome-mining predicted it to encode 52 BGCs for natural compound syntheses.
For subsequent analyses, they mostly focused on BGCs related to the production of polyketides (PKs) and non-ribosomal peptides (NRPs), but reported data for the other predicted types of compounds as well.
Time-course experiment
“We conducted a time-course experiment of the myxobacterium Sorangium sp. So ce836 in a liquid batch culture, over five to six time points, covering all growth phases,” said first author Dr. Judith Boldt.
“From this, we obtained time-resolved transcriptomics (RNA-seq) and metabolomics (LC/MS) data, and we put the transcription of BGCs into the context of genome-wide transcription and molecular function.”
The team found that the transcription levels of core biosynthetic genes of most BGCs were actively transcribed at specific time points. The majority (80%) of PKS and NRPS genes belonged to the transcription groups with maximal transcription in the exponential phase.
Linking the time-course transcriptomics data of BGCs with the time-course metabolomics data of the corresponding known natural compounds (specifically icumazole A and epothilone A), they found that the time point of maximal transcription coincided with the time point of maximal net production rate per cell of the compound, indicating a regulation at the transcriptional level for these compounds.
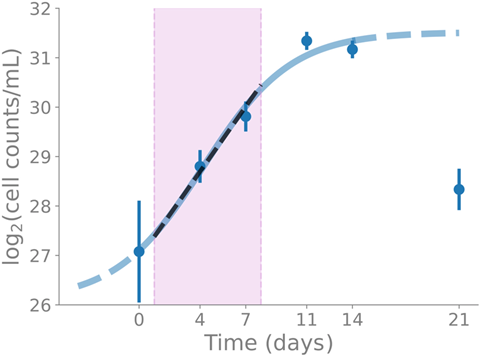
“We have shown that the transcription of BGCs in Sorangium sp. So ce836 is determined by bacterial growth, shedding light on the regulation of BGC transcription,” Dr. Boldt said.
“The majority of BGCs in the genome were transcriptionally active, contradicting the assumption of widespread transcriptional silence of BGCs in laboratory cultures.
Widening the search
“Together with our observation of maximal transcription levels and natural compound net production rates in growing myxobacterial cells (exponential phase) this indicates that studies of natural compounds (especially in myxobacteria) should not be limited to nutrient-limited conditions or the stationary growth phase.
“It suggests that adjusted batch runtimes, production retention approaches, or even continuous fermentation might offer the potential for maximisation of biotechnological product yields.”
The temporal dynamics of BGC transcription, translation, and natural compound assembly in bacteria are poorly understood, she said, advising that time-course transcriptomics, proteomics, and metabolomics studies of other organisms are needed to assess if these findings are specific to this one organism or more widely generalizable.
Finding out more
“Improved genetic engineering tools for So ce836 or a related Sorangium sp. strain would facilitate more detailed analyses and manipulation of transcriptional regulation of their BGCs, the natural compounds of which in most cases are not yet known.
“A more detailed understanding of BGC expression and regulation will facilitate the discovery of novel, biotechnologically useful natural compounds, the triggering of their biosynthesis, and the optimization of their large-scale production.”
This study was a collaboration between the Leibniz Institute DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) and the Helmholtz Centre for Infection Research. The work was partially funded by the German Center of Infection Research (DZIF) and by the Leibniz Institute DSMZ.
‘Bursts in biosynthetic gene cluster transcription are accompanied by surges of natural compound production in the myxobacterium Sorangium sp.’ appears in Microbial Biotechnology.






No comments yet