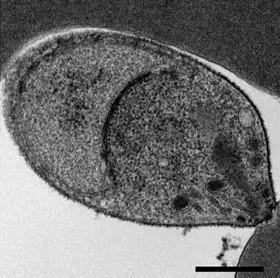
A new, comprehensive map of all the genes essential for blood infections in Plasmodium knowlesi (P. knowlesi), a parasite that causes malaria in humans, has been generated by researchers at Harvard T.H. Chan School of Public Health and colleagues. The map contains the most complete classification of essential genes in any Plasmodium species and can be used to identify druggable parasite targets and mechanisms of drug resistance that can inform the development of new treatments for malaria.
“We hope that our findings are a major step for the field of malaria research and control,” said co-corresponding author Manoj Duraisingh, John LaPorte Given Professor of Immunology and Infectious Diseases. “Emerging drug resistance to the small number of antimalarial drugs is a growing problem. This map will be an invaluable resource to help researchers combat one of the leading causes of infectious disease death around the world.”
The study will be published Feb. 6, 2025, in Science.
Zoonotic parasite
Each year, around 249 million human cases of malaria are caused by a Plasmodium species, resulting in about 608,000 deaths. P. knowlesi is one of several species responsible for human malaria. It is a zoonotic parasite that can be lethal and is an emerging public health problem in Southeast Asia.
READ MORE: Scientists design new drug to fight malaria
READ MORE: Scientists identify molecules associated with recurrence in blood samples from malaria patients
The researchers developed a powerful genetic approach in P. knowlesi, known as transposon mutagenesis, to disrupt all of the genes not needed for growth in human red blood cells, thus revealing a map of all of those remaining that were essential for growth. This identified at a genomic scale the molecular requirements for parasites to grow. The researchers were also able to pinpoint specific genes that cause resistance to current antimalarials.
“Knowing all of the essential genes in P. knowlesi allows us to understand the molecular strategies that the parasite takes to grow, to respond to environmental changes, and to respond to therapeutics such as antimalarials,” said co-first author Sheena Dass, postdoctoral fellow in the Department of Immunology and Infectious Diseases. “This molecular blueprint will help malaria researchers in the design and execution of biological studies of malaria as well as inform strategies to monitor and limit the emergence of drug resistance.”
Insight into sister species
The researchers noted that the findings also provide insights into one other malaria-causing Plasmodium species, P. vivax, due to their evolutionary relatedness. P. vivax, which is poorly studied because it is not culturable or genetically tractable, is a major challenge to elimination efforts.
Brendan Elsworth and Sida Ye were co-first authors. Co-corresponding author was Kourosh Zarringhalam at the University of Massachusetts, Boston.
Other Harvard Chan co-authors included Jacob Tennessen, Basil Thommen, Aditya Paul, Usheer Kanjee, and Christof Grüring.
The study was supported by the National Institutes of Health (grants 5R01AI168163, 5R01 AI167570, ORIP/OD P51OD011132, and U42 PDP11023); the Swiss National Science Foundation Postdoc Mobility (fellowships PBSKP3_140144 and P300P3_151146); and the Food and Drug Administration Intramural Research Program.
Topics
- Aditya Paul
- Basil Thommen
- Brendan Elsworth
- Christof Grüring
- Harvard T.H. Chan School of Public Health
- Jacob Tennessen
- Kourosh Zarringhalam
- malaria
- Manoj Duraisingh
- Microbial Genetics
- One Health
- Parasites
- Plasmodium knowlesi
- Research News
- Sheena Dass
- Sida Ye
- University of Massachusetts, Boston
- USA & Canada
- Usheer Kanjee
- Veterinary Medicine & Zoonoses




No comments yet