It’s an unpleasant fact that most of us are happy to ignore: Our mouths and noses are the natural homes to infectious and antibiotic resistant bacteria.
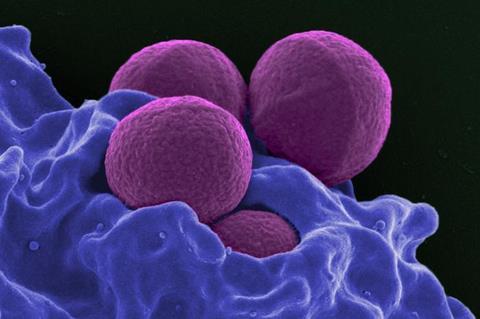
The good news is that our nasal and oral environments are usually equipped to keep these germs in check, said Gemma Reguera, a professor in the Department of Microbiology, Genetics and Immunology at Michigan State University.
READ MORE: Thyme essential oil inhibits staph biofilm formation
READ MORE: Staph mechanism for resistance to last resort drug is uncovered
It’s when these microbes, such as Staphylococcus aureus and its antibiotic-resistant variant MRSA, move deeper into the airways or into other body sites, like the heart, that they can cause serious, even fatal, infections.
Highway to the danger zone
Reguera and her colleagues have now shown that our bodies can unwittingly create conditions that usher staph, including MRSA, right into those danger zones.
“You know the people who wave the sticks to direct planes at the airport? It’s like that,” said Reguera, whose team published the new research in the journal mBio.
The flip side is that this report also provides new insights and techniques that can help researchers prevent our natural inhabitants from spreading to places they shouldn’t.
“These are organisms that we harbor,” Reguera said. “It’s like having neighbors you don’t like. We need to learn to live with them.”
Staphylococcus slide
Understanding how these bacteria move into different parts of the body is a complex puzzle underscored by two simple facts.
One, the microbes obviously move around and make people sick. Two, they aren’t well equipped to do so. Staph, MRSA and their ilk lack the appendages, such as tail-like flagella, that help other types of bacteria get around.
Confronting these two seemingly conflicting facts is a problem that’s new to Reguera’s group.
She and her team are perhaps best known for their extensive work with soil bacteria that they’ve shown can do things like sop up radioactive elements. But, with support from the Office of Naval Research, the researchers have brought a unique ecological perspective to the studies of staphylococci — the plural of staphylococcus — and other bacteria inhabiting our noses, mouths and middle ear.
Microbial ecology
“What’s fascinating to me about the perioral environment is how environmentally diverse it is and how much its chemistry can change within such a small space,” Reguera said. “This was new territory for me and a huge intellectual effort because of the impressive body of literature about human-associated bacteria. But we did what we do best: We looked at these microbes through the lens of microbial ecology.”
Kristin Jacob, who worked on this project as a doctoral student, and Santiago Hernandez-Villamizar, a visiting Ph.D. student from the University of Los Andes in Colombia, helped forge the lab’s path into this new realm.
The team first discovered that certain proteins called mucins made by our mucous membranes acted as lubricants for colonies of staphylococcus bacteria. These mucins made it easier for the microbes to spread, but it still wasn’t enough to explain how well they got around in worst-case scenarios.
Active and coordinated
“We found that the colonies can passively expand when there’s enough lubrication,” Reguera said. “But you need more than something passive to enable an infection. That’s much more active and coordinated.”
By culturing different strains of various species of staphylococcus isolated from healthy human volunteers, the team systematically cultivated a pool of contestants with diverse genetics to see which could actively spread in a simulated biological environment.
They found that, in the right conditions, the winners were Staphylococcus aureus and a species known as Staphylococcus epidermidis. When colonies of these bacteria grew large enough, the microbes in the crowded periphery would signal to each other that it was time to make their own lubricants and slide away fast.
Crowd-sensing system
To dig deeper, the group teamed up with Neal Hammer, an associate professor at MSU and an expert in staphylococci. The team worked with MRSA strains that had certain parts of their crowd-sensing systems deactivated. In doing so, they identified peptides that the bacteria were making that also acted as lubricants to aid their movement.
The right combination of mucins and secreted peptides threw open the doors, almost literally, to the spread of the bacteria.
“We basically have chemicals in our mucus surfaces that are like rolled out red carpet for these bacteria toward our most vulnerable sites,” Reguera said. “All of a sudden, the bacteria start making their own peptides and they’re unstoppable. They can colonize new areas and grow.”
Host of new questions
Although this discovery begets a host of new questions that need to be answered before providing new therapeutic ideas, it also provides new tests and techniques to aid researchers in that quest.
“The clinical and therapeutic aspects of this work will really drive how we look at this phenomenon going forward,” Reguera said.
In the meantime, Reguera is enjoying a sense of personal and professional accomplishment with her lab’s growth into a new research area. She credits that success to the teamwork of industrious and fearless graduate students, as well as a generous and collaborative co-investigator in Hammer.
“Never in my whole life did I think I’d work with staphylococci,” she said. “I’m very proud of my students and grateful to my colleague and now collaborator, Neal Hammer, for his expert insights.”







No comments yet