Salmonella and E. coli are well-known bacteria that cause food poisoning, but less understood are species of Providencia, another causative agent of serious symptoms.
Providencia rustigianii, isolated from pediatric gastroenteritis patients, has now undergone whole genome sequencing by a research team led by Osaka Metropolitan University Professor Shinji Yamasaki of the Graduate School of Veterinary Science and the Osaka International Research Center for Infectious Diseases.
READ MORE: Wastewater monitoring can detect foodborne illness
READ MORE: Spread of diarrheal illness linked to climate change
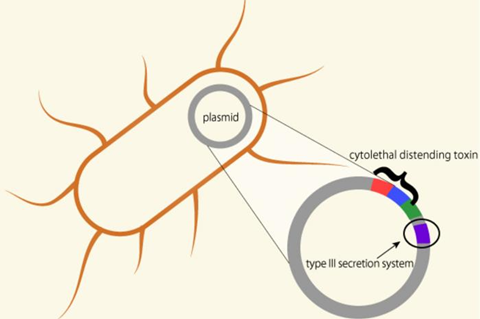
Members of the team had previously reported that P. rustigianii carries a cytolethal distending toxin virulence gene on its plasmid. Elimination of this gene did not reduce the bacteria’s virulence, however.
Virulence factor
This time, the whole genome sequencing revealed that P. rustigianii also possesses a virulence factor called the type III secretion system, which bears the highest degree of similarity to the gene found in Salmonella. The type III secretion system was found to be directly involved in cell invasiveness and enterotoxicity, making it a major virulence factor for P. rustigianii.
“Using the pathogenicity gene discovered in this study as an indicator, we plan to create a detection system for pathogenic species of Providencia in wild animals, water, livestock, and food, which can be expected to be useful in investigating natural hosts, sources of infection, and infection routes,” Professor Yamasaki explained.
“In addition, clarification of the pathogenic mechanism by the type III secretion system is expected to lead to the development of therapies that do not use antimicrobial agents.”
The findings were published in mBio.






No comments yet