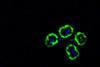
Bacteria colonise different habitats in complex communities, including in the human gut or on the skin. Analysing these bacterial communities - the microbiome - is an extremely complex task. A central problem in microbiome analysis is the so-called “bias” that occurs in the various steps of microbiome research, from sample collection to sample analysis.

These bias effects are particularly challenging during extraction, i.e. the extraction of bacterial DNA. Different extraction methods can obtain the DNA of certain bacterial species differently, which can significantly distort the determined microbiome composition. This jeopardises the accuracy of important scientific and clinical findings.
Computer-based method
Scientists at the Department of Environmental Medicine at the University of Augsburg have now developed standardised methods to address this problem. In a recent study, Dr Luise Rauer from the team of Prof Avidan Neumann, head of the Department of Environmental Bioinformatics, has developed an innovative, computer-based method for correcting the extraction bias.
READ MORE: Temper the hype of human microbiome studies
READ MORE: Report recommends ‘highly ambitious enterprise’ to create world-leading UK Microbiome Biobank
“The method is based on the general morphological properties of the bacteria, for example the shape of their cells and the stability of their cell walls. This means we can apply it regardless of the extraction method used and the specific type of bacteria. In principle, it can be used for all types of microbiome samples, for example gut, skin or environmental samples,” explains Luise Rauer.
Mock communities
In the study, the researchers compared microbiome samples from skin samples and so-called mock communities, i.e. artificial bacterial communities with a known composition, using different extraction methods. The distortion effects of the extraction were successfully reduced by the new calculation method, which led to a significantly more accurate determination of the composition of the microbiome.
“Microbiome tests help to better understand diseases. The results of this study could significantly improve microbiome analysis in clinical applications such as diagnostics and personalised medicine. More reliable and consistent results can be crucial in clinical practice,” adds Prof Dr Claudia Traidl-Hoffmann, Chair of Environmental Medicine at the University of Augsburg and Director of the University Outpatient Clinic for Environmental Medicine at the University Hospital.




No comments yet