This study, published in mLife, was led by Profs. Chang‐Fu Tian and Ziding Zhang (College of Biological Sciences, China Agricultural University).
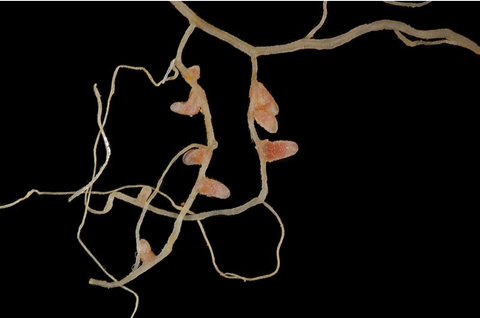
Based on a robust Tn-seq analysis of independent mariner transposon insertion libraries of Sinorhizobium strains, pangenomic and network-based analyses allowed identification of a strain-dependent variation in the fitness network (harboring ES, GA, GD, and NE genes) of Sinorhizobium pangenome under a nutrient-rich condition.
This fitness network is characterized by a highly connected ES subnetwork, and beneficial (GA) and deleterious (GD) subnetworks of lower connectivity.
Genus core genes belonging to both the shared and strain-dependent essential zones of this fitness network exhibited a similar profile of functional categories, e.g., cell envelop biogenesis (Figure 1).
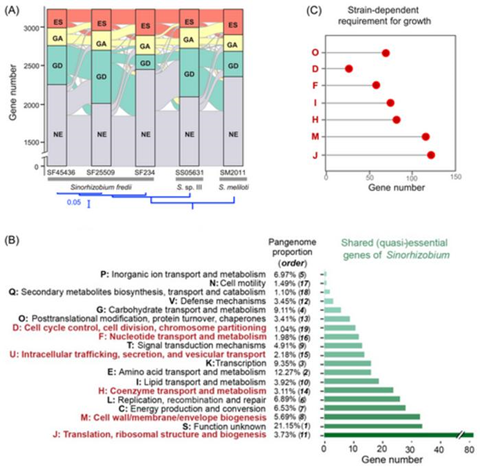
The network-based analyses of the fuzzy essential zone of Sinorhizobium pangenome developed in this work can be used for any prokaryotes for which a robust Tn-seq procedure can be established. These efforts are significant for fully understanding the evolution of prokaryote pangenome, the in silico bipartition of which into essential core and non-essential accessory subsets is oversimplified.






No comments yet