Light and electron microscopy have distinct limitations. Light microscopy makes it difficult to resolve smaller and smaller features, and electron microscopy resolves small structures, but samples must be meticulously prepared, killing any live specimens.
Atomic force microscopy (AFM) is a technique originally developed to assess the physical and mechanical properties of materials at extremely high resolutions, but the imaging speeds aren’t fast enough (e.g. several minutes per frame) to capture relevant data for living biological samples. In contrast, another method, high-speed AFM (HS-AFM), is fast but cannot measure mechanical properties.
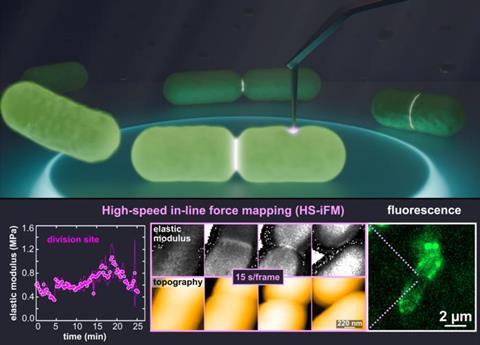
Understanding the potential of this type of microscopy for the analysis of large molecules and microorganisms, researchers from the National Institutes of Natural Sciences (NINS) and Nagoya University created a new technique, high-speed in-line force mapping (HS-iFM), to acquire dynamic, mechanical force measurements at the speed and resolution required for living biological samples. The group – Christian Ganser, Shigetaka Nishiguchi, Feng-Yueh Chan, and Takayuki Uchihashi – chose the common bacteria Escherichia coli to study first in such unprecedented detail.
The researchers published their study in the January 29 issue of Science Advances.
Living organisms
“While it is clearly informative to study ‘static’ samples, [such as] bacteria that are not alive, observing the living organism allows us to directly follow changes in the course of [an organism’s] life, which becomes possible with our technique. Escherichia coli are great to demonstrate such effects because they are well studied. Despite this, the dynamic [mechanical] changes [that occur in E. coli at the] nanoscale level… remain elusive,” said Christian Ganser, assistant professor at the Exploratory Research Center on Life and Living Systems (ExCELLS) in the NINS in Okazaki, Japan.
READ MORE: Breakthrough in nanotechnology: Viewing the invisible with advanced microscopy
READ MORE: High speed atomic force microscopy studies provide insights into influenza A viral replication
During E. coli cell division, the researchers observed increased mechanical stiffening of the cell division site. The researchers hypothesize this stiffening could be due to localized membrane tension and local cell wall thickening. “The division site becomes much stiffer than the surrounding cell, hinting at large internal stresses that are needed to deform the membrane and separate the cells,” said Ganser.
Visible bridges
During division, the membrane also formed visible bridges between the two daughter cells that stretched and eventually broke. The creation and eventual breakage of these bridges lasted an average of 242 s ± 99 s (mean ± standard deviation) and was observed seven times. The entire division process is provided as a supplementary video in the online version of the research paper.
The team also observed a weak spot, less than 100 nm in diameter, in a dividing E. coli cell that ruptured, causing cell depressurization and death. Interestingly, the bursting cell caused depressurization of both left and right daughter cells, indicating that the cells were not completely separated internally. This observation suggests that HS-iFM may be useful in determining the timing of different steps during the division of E. coli and other bacteria.
Membrane holes
HS-iFM allows researchers to measure both high resolution topography and membrane mechanical properties. During division of a living E. coli cell, the researchers observed visible membrane holes that close and reform again and diffuse across the membrane. The team hypothesized that these dynamic hole structures may be related to the formation of outer membrane vesicles, which are reported to be more frequent during cell division and around the new cell walls that develop between daughter cells during bacterial cell division. The pores could also be outer-membrane protein complexes located on the bacteria surface, but the measured pore diameter of 34.7 nm ± 11.8 nm (1 nm = 1 x 10-9 m) is significantly larger than previous reports of protein complex diameter, which is approximately 8 nm.
The research team acknowledges the great potential of the HS-iFM technique for studying a wide variety of biological samples and organisms, including E. coli. “In the [future], we will use our technique to study the dynamic and localized effect of external stimuli, [such as] antibiotics, to the nanomechanical properties of membranes of living bacteria,” said Ganser. The researchers also envision using HS-iFM to study the transient nanomechanical properties of polymers. Ideally, the team will increase the speed and resolution of the technique to visually capture the mechanical properties of molecules as small as individual proteins.
The co-authors of the research paper are Shigetaka Nishiguchi from the Exploratory Research Center on Life and Living Systems at the National Institutes of Natural Sciences in Okazaki, Japan; Feng-Yueh Chan from the Department of Physics at Nagoya University in Nagoya, Japan; and Takayuki Uchihashi from the Exploratory Research Center on Life and Living Systems at the National Institutes of Natural Sciences and the Department of Physics at Nagoya University.
This research was supported by JSPS KAKENHI Grant Number JP23H04874 in a Grant-in-Aid for Transformative Research Areas “Materials Science of Meso-Hierarchy” and 24K01309, 22K18943; by JST, CREST Grant Number JPMJCR21L2, Japan; and MEXT Promotion of Development of a Joint Usage/Research System Project: Coalition of Universities for Research Excellence Program (CURE) Grant Number JPMXP1323015482.




No comments yet