Unicellular microbes are certainly the oldest life forms on Earth. They may have appeared around 3.5 billion years ago and have played essential roles in shaping the biosphere as we know it today.
How they evolved and diversified over time remains, however, largely a mystery. Most microbes do not produce mineralized structures that facilitate fossilization, and their minute size, hard-to-distinguish forms, and structures do not allow for easy identification or the taxonomic assignment of fossilized remains.
Is there really no way to objectively address the history of microorganisms?
The answer is (to some extent) “Yes there is”, and the keyword is paleogenetics.
Who is not today familiar with images of crime scenes where detectives search for traces of DNA left behind by murderers? What this quest tells us is that traces of our physical presence survive, long after we are gone. This property, which holds true for all living creatures including microbes, is at the heart of paleogenetics. This discipline studies the degraded DNA extracted from the remains of ancient living beings to highlight their evolution and relationships to extant species and individuals.
Paleogenetics emerged 40 years ago with the characterization of DNA sequences extracted from a quagga museum specimen, a zebra species that went extinct at the end of the 19th century. While this research field has been popularized for its contribution to human evolution with the 2022 Nobel Prize in medicine awarded to Svante Pääbo for the sequencing of the Neanderthal genome, it is swiftly expanding across the tree of life and has, as we will see, already made major advancements in microbiology.

There is, however, at present, a time limit to paleogenetics. Indeed, DNA does not survive indefinitely in the environment and not all environments are favorable to its permanence. As of 2023, the oldest environmental sample from which DNA has successfully been characterized is a 2 million-year-old frozen sediment collected in northern Greenland, which encapsulated the plant and animal diversity that characterized the corresponding Early Pleistocene landscape.
Paleomicrobiology and ancient DNA
Thus, paleogenetics, and more specifically molecular paleomicrobiology, is bringing microorganisms from the past to light, including in some special cases their entire genomic sequences. Sources of ancient microbial DNA are extremely diverse; they range from environmental samples such as freshwater or marine sediment cores; archaeological artifacts such as long-buried animal and human skeletons; or more recent items stored in natural history collections (zoological collections, herbaria). Ancient DNA (aDNA) is however often difficult to access, limited in quantity, and degraded. Its extraction and manipulation require the implementation of specific protocols and procedures. aDNA extractions are usually performed in dedicated pressurized clean rooms where airborne particles are maintained at a very low concentration to minimize contamination by modern genetic material. aDNA also differs from modern DNA by the presence of specific chemical and physical modifications that spontaneously arise over time. The two main postmortem damages (PMD) observed in aDNA molecules are depurination which leads to extreme DNA fragmentation, and deamination, which instead leads to the substitution of specific nucleotides, most commonly at the extremities of the DNA fragments. Quantification of these modifications is important in order to confirm the ancient origin of the extracted DNA.

Evolution of past microbial pathogens
Molecular paleomicrobiology is now making essential and original contributions to the field of medical microbiology by addressing key questions that cannot be answered using extant strains of pathogens. These key questions regard the emergence and spread of pathogens.
Have they always been associated with humans? How did virulence evolve over time?
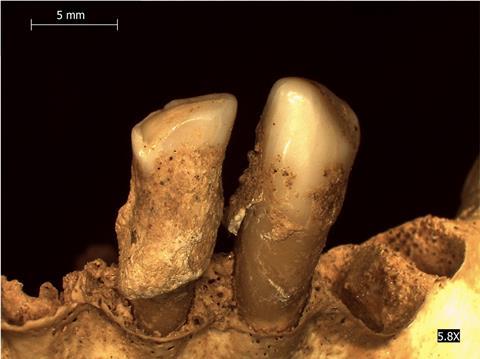
In this respect, the many studies performed on Yersinia pestis, the causal agent of plague, are particularly illustrative. Since septicemia usually precedes death, DNA from this pathogen is usually recovered from the highly vascularized dental pulp of skeletons found in mass graves characteristic of plague epidemics. Its presence in skeletons unearthed from graves that date back to the historical “Justinian” (540-770 CE) and “Black Death” (1346-1351 CE) pandemics confirmed that it was indeed the culprit. Besides these documented epidemics, the presence of Y. pestis DNA in human remains from as far back as the bronze age (5000-3500 years ago) points to a longer than previously suspected interaction between humans and this bacterium. Finally, reconstruction of entire historical genomes of this species has allowed dating the emergence of key genetic features known to be important for its transmission and virulence. It was thus shown that a majority of the most ancient (bronze age) genomes lacked several of the genes or mutations essential for multiplication and maintenance in the intermediate insect (flea) host. These ancient strains may thus not have been transmitted to humans through insect bites and, consequently, may not have provoked the emblematic and highly contagious bubonic form of the disease.
While the study of the causal agent of plague has dominated the landscape of paleomicrobiology, similar scientific studies have been pursued for other pathogens, including those that infect plants. These studies are not limited to bacterial pathogens (e.g. Mycobacterium sp., Xanthomonas citri), but also illustrate the historical spread and adaptation of viruses (e.g. Hepatitis B virus, smallpox virus), or of eukaryotic parasites, as in the case of the Oomycete Phytophthora infestans that was responsible for the devastating potato blight disease and great potato famine in Ireland.
From single pathogens to complex microbial communities
Paleomicrobiology is however not restricted to the study of single individual microbial species. The approach extends also to the study of entire and diversified microbial communities such as those associated with macroorganisms (humans, animals, or plants). These host-associated microbial communities can be affected by their host’s identity (species, genotype), lifestyle (diet, health status), and local environment. Several paleogenomic studies have thus tried to disentangle the relative importance of the host’s status, geography, ecology, and environment in the temporal evolution of their microbiomes, all of which contribute to the fitness and health of their host and their selective value.
Not all the host-associated microbial communities’ DNA survives equally throughout time. Feces, for example, whose content reflects the gut microbiome, are only preserved in exceptional instances, such as in salt-rich (mines) or desert environments that allow for a rapid desiccation of the samples. Numerous published studies on host-associated microbiomes focus instead on mammalian (including human) oral microbiomes. Indeed, mineralization of the dental plaque produces dental tartar (calculus) which encapsulates the microorganisms present and preserves their DNA for long periods. An illustrative and original example of the use of this material in molecular paleomicrobiology regards the analysis of dental calculus from Swedish wild brown bears that died between 1842 and 2016 and whose skulls were stored in natural history collections. This study coincidentally revealed a posteriori the impact of regulatory policies for the use of antibiotics on the prevalence of antimicrobial resistance in the environment. While antibiotics were not available before 1951, the period 1951-1985 corresponded to their widespread use in humans and animal husbandry. After that date, measures were implemented in Sweden to reduce their use, including their ban as animal growth promoters. Molecular analysis of aDNA extracted from the bears’ dental calculus revealed that the abundance and also the diversity of bacterial antibiotic resistance genes changed significantly over time in the corresponding oral microbial communities. Trajectories of these changes reflected both the different phases of antibiotic use in Sweden and their widespread impact even on wild animal species not closely associated with humans and farm animals.
Similar studies were conducted on the oral and intestinal communities of ancient and modern human populations that differed with respect to diet and lifestyles (e.g. Neolithic hunter-gatherers versus farmers). Both the studies of ancient human oral and gut microbiomes concluded that there was an increase in the prevalence of antibiotic resistance genes in the microbiomes of extant human populations (from either industrialized or non-industrialized societies), compared with those in ancestral populations.
In the case of plants, a much more limited number of studies have examined their microbiomes using DNA extracted from either the aboveground (leaves, stems) or underground (roots) organs of dried herbarium specimens. Herbaria have mostly been established in the 19th and first half of the 20th century, a period that precedes today’s acceleration of global changes whose impacts on the earth’s microbiome are poorly documented. Analysis of microbial aDNA extracted from herbarium plants, as well as from ancient soil that is sometimes still present around their roots should inform us on how, and to what extent global changes have durably affected not only plant microbiomes but also the soil.

Future perspectives
Molecular paleomicrobiology is revolutionizing microbiology by inserting microorganisms into a historical framework – something classical microbiology could not do. This groundbreaking discipline not only unveils the mysteries of the past evolution of microorganisms but also serves as an entry point to exploring uncharted territories of the microbial realm. In this respect, the de novo assembly of genomes from microbes in dental calculus dating back to 100,000 years ago, has unveiled species previously unknown to science and represents a new milestone for the discipline.
One particularly fascinating result regards the discovery of a secondary metabolite biosynthetic gene cluster present in the oral microbiomes of both Neanderthals and modern humans who lived during the Middle and Upper Paleolithic periods. This cluster, present in the genome of a bacterial species affiliated with the genus Chlorobium, was synthesized in vitro as a full-length DNA molecule and was transferred into the modern bacterium Pseudomonas protegens for expression. This ingenious approach led to the characterization of two novel molecules, named paleofuran A and B, which expand the catalog of known bacterial secondary metabolites.
Beyond its immediate impact on microbiology, this proof-of-concept approach that resuscitates ancient metabolic pathways and their products may have broader implications in biotechnology. This approach indeed paves the way for the direct study of the evolution of microbial natural products and for the potential recovery of bioactive substances that extant microbes may no longer produce. Screening of these molecules for biological activity may lead to the identification of new drugs that could be used in medicine.
The authors’ upcoming paper entitled ’Paleomicrobiology: tracking the past microbial life from single species to entire microbial communities’ has been accepted for publication in Microbial Biotechnology.
Further Reading
Ancient DNA analysis | Nature Reviews Methods Primers
Natural products from reconstructed bacterial genomes of the Middle and Upper Paleolithic | Science













No comments yet