Biological membranes separate cells and organelles from their external environment. Composed of a thin (~6 nm) bilayer of lipids with embedded and bound proteins, membranes and their components are involved in almost all cellular processes. The importance of biological membranes is such that ~60% of drugs target membrane components.
Over a thousand different lipids are found across all known life forms and over eight thousand differing membrane proteins are found in humans alone, making biological membranes highly complex in terms of their protein and lipid constituents.
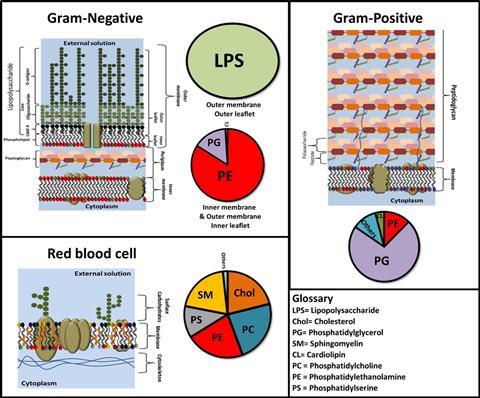
Membrane architecture differs across differing cell types. Consider Gram-negative bacteria like Escherichia coli, whose outer membrane displays a distinctive asymmetric lipid arrangement. This outermost membrane possesses an inner leaflet comprised of phospholipids that face the cell, while its outer leaflet, exposed externally, consists of lipopolysaccharides – a bacteria-specific molecule. These lipopolysaccharides feature extended sugar chains oriented away from the membrane’s lipid core, serving as a defensive mechanism against the external environment for the bacteria.
In contrast, mammalian cells exhibit a membrane primarily composed of phospholipids and sphingolipids, enriched with a substantial cholesterol content that finely regulates its fluidity.
The spectrum of membrane proteins displays remarkable diversity across the kingdoms of life. Numerous proteins, both integral and peripheral, play crucial roles in a vast array of biochemical functions, such as cellular signaling as well as the intake and export of nutrients and materials into and out of the cell.
Model membranes
The thin width and structural complexity of biological membranes make gaining a structural understanding of membrane biochemical events with molecular precision challenging when using live cells. To fill this knowledge gap, artificial biological membranes have been developed. These models have a reduced complexity compared to “real” biological membranes. Examples of model membranes include lipid vesicles which are hollow spheres of solution enclosed by a lipid bilayer, and supported lipid bilayers which are flat model membranes deposited onto a support material.
Supported lipid bilayers have been extensively used to gain a precise understanding of membrane-related phenomena due to their ease of fabrication and the number of surface analysis techniques that can be utilized to probe these samples. One technique that is gaining increased use in the analysis of the structure across supporting lipid bilayers is neutron reflectometry.
Neutron Scattering in Biology
The existence of the neutron had been theorized by Ernest Rutherford in his model of the atom but wasn’t proven until experiments undertaken by James Chadwick in 1932. Research involving neutrons was initially focused mainly on energy and military applications. As a by-product of this, the use of neutron beams as a tool for examining the atomic and molecular structure of materials was developed. By the 1970s, dedicated research facilities began operation, which used neutron scattering techniques to unravel the structure and dynamics of materials.
The benefit of neutron scattering in the study of biological structures was realized around this time. It had long been understood that the neutron scattering magnitude (known as the scattering length) of protium – the hydrogen isotope which makes up 99.98% of the hydrogen found on earth – and deuterium (which constitutes the other 0.02%) are highly differing in value. This means that the neutron scattering length density (which is related to the refractive index) of water (H2O) and heavy water (D2O) have values that are essentially opposite.
Neutron scattering from a molecule derives from a difference in its neutron scattering length density compared to its surrounding environment. Biological macromolecules such as proteins, ribonucleic acids, lipids, and carbohydrates, all have different neutron scattering length densities falling between those of H2O and D2O. This unique characteristic gives neutrons a distinct advantage in examining bio-macromolecular complexes. When examining a biological complex – an assembly containing two or more macromolecules – the H2O:D2O ratio around the complex can be modified to either amplify or diminish the scattering signal from individual components. This means that neutron scattering can uniquely resolve the relative distribution of structural components in biological macromolecular complexes. This capability has been particularly exploited to gain a precise understanding of membrane biochemistry.
Neutron Reflectometry and Model Membranes
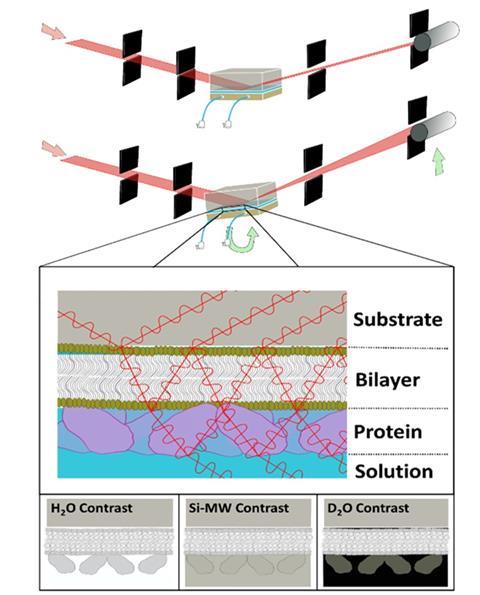
There are several neutron scattering techniques that are used to study biological membranes. One technique that is commonly utilized to examine membranes is neutron reflectometry. This technique measures structures across interfaces through the interference between neutrons reflected at differing scattering length density boundaries (akin to refractive index). Originally developed for applications in chemistry and physics, it has recently been extensively employed to measure membrane biochemical events modeled with supported lipid bilayers. In this approach, a model membrane is deposited either onto or adjacent to a surface, and neutron reflectometry is used to resolve its structure before and after a membrane biochemical event.
Like all neutron scattering techniques, hydrogen/deuterium labeling around the sample can be utilized to resolve the relative structural distribution of components within the samples. As neutrons are a non-damaging structural probe, neutron reflectometry can be used to measure the structure of a model biological membrane before, during, and after a membrane biochemical event, such as peripheral membrane protein binding, the disruptive effects of antimicrobials to pathogen membranes, and the effects of toxins on mammalian and plant cellular membranes.
Antimicrobial Interactions
A common area of use of neutron reflectometry is to examine the disruptive effects of membrane-perturbing antibiotics on model pathogen membranes. These models can include both Gram-negative, Gram-positive, and fungal pathogen models. In these studies, a model of the pathogen’s membrane is fabricated onto a planar surface – usually a substrate such as silicon or quartz – and the structure across the membrane is then resolved with neutron reflectometry. The antibiotic, toxin, or protein under study is then added to the solution medium adjacent to the model membrane, and both its effect on the structure of the membrane and its position relative to the membrane is structurally resolved. This essentially provides a precise method for benchmarking activity.
There are many examples of this use including examining the mechanisms of both existing and new peptide antibiotics, as well as the disruptive effects of proteins that make up the innate (non-adaptive) immune system of animals and plants. One such example of the latter is work conducted examining the antibacterial mechanism of lactoferrin, a large dumbbell-shaped protein found in mammalian milk that is known to specifically kill Gram-negative bacteria. It was found that the large protein was able to bind end-on to the bacterial outer membrane surface through a specific interaction with the lipopolysaccharides on the membrane surface and caused a loss of membrane integrity, with a large increase in water in the membrane.
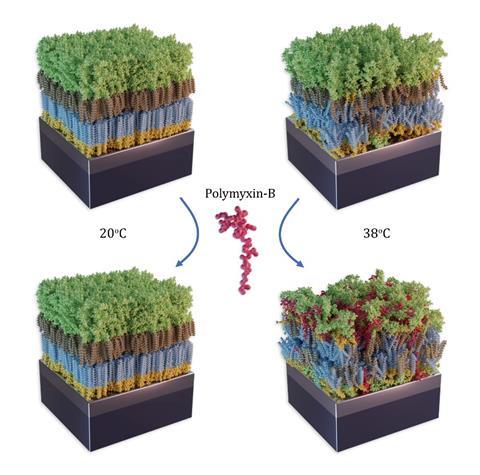
Polymyxin B is a cyclic lipopeptide-based antibiotic first approved for use as a clinical antibiotic in 1964. Like lactoferrin, it specifically targets the lipopolysaccharides found in the outer leaflet of Gram-negative bacteria’s outer membrane. Using neutron reflectometry, the details of the drug’s ability to disrupt the bacterial outer leaflet were uncovered. Most notably the fluidity of the lipopolysaccharide in the outer membrane is key to the drug’s ability to penetrate the membrane and disrupt it.
Understanding the relationship between pathogen membrane dynamics and antimicrobial activity provides important information not only in understanding those currently used to treat infections but also aids in the rational design of new antibiotics to help combat the growing threat of antimicrobial resistance.
Toxin Interactions
Similar to studies on antimicrobial disruption to membranes, neutron reflectometry can also be utilized to examine how toxins disrupt animal and plant cellular membranes in their toxic activity. Examples of this work include elucidating the membrane disruptive activity of snake venom.
Recently, work was conducted looking at a toxin produced by oomycetes – single cellular eukaryotic plant pathogens found in soil. These microorganisms are of interest due to their ability to cause blight, a crop-destroying plant disease that has been historically associated with famines. In their plant pathogenic activity, oomycetes release toxins that destroy plant cells. Using neutron reflectometry combined with other techniques, the disruptive mechanism of one such toxin – NLP1 (necrosis-like protein 1) – was uncovered. Data revealed that the protein toxin was only able to interact with membranes containing a particular lipid type found only in plant membranes (Glycosyl Inositol Phospho Ceramides) when it bound to the membrane surface, inducing small pores. This mechanism is novel, and now it has been characterized that this information can be utilized in the design of environmentally safer pesticides.
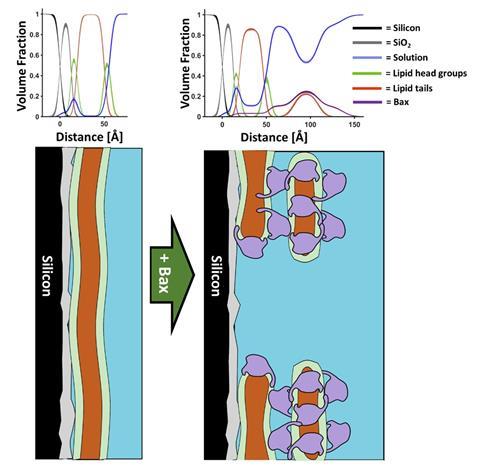
Another recent example of an important membrane biochemical event that was resolved using neutron reflectometry was the initial stages of programmed cell death known as apoptosis. Apoptosis is a vital cellular process that removes diseased and damaged cells, preventing cancer. In a human adult, this process occurs between fifty to seventy billion times a day. Apoptosis is initiated at the mitochondrial membrane surface where a protein called bax is sequestered to the outer membrane of this double membrane organelle. This causes pores in the outer membrane which release a series of cell death factors into the cell cytoplasm – which ultimately leads to cell death. Neutron reflectometry provided precision structural insight into the mechanism by which bax disrupted the mitochondrial outer membrane, showing that the protein was able to form pores by removing lipids from the membrane and depositing them onto the mitochondrial surface in the form of bax-lipid clusters. This work revealed that the formation of the membrane pores and the clusters was proportional, showing how bax was essentially able to restructure the mitochondrial outer surface in the initial stages of apoptosis. Understanding how important mammalian proteins such as bax function with such precision allows for not only a greater understanding of our biochemistry but also provides information that can be used in the design of new therapeutics such as anti-cancer drugs.
Future Applications and Conclusions
As neutron reflectometry continues to develop, its applications are expanding to the investigation of more sophisticated membrane biochemical events. In particular, there is a growing focus on using this technique to examine membrane-spanning proteins within their native lipid matrix, due to the importance of integral membrane proteins in a wide range of biochemical and pharmaceutical phenomena. Advancements in this technique also hold promise in contributing to our understanding of antimicrobial resistance, a pressing global concern in the field of healthcare and pharmaceuticals.









No comments yet