The fascinating world of bacteria that live as symbionts or parasites in animal hosts often remains a mystery to researchers. Kiel University (CAU) and the Max Planck Institute for Marine Microbiology in Bremen are contributing to solving this puzzle with their research into the interactions between microbes and their host.
Under the leadership of Prof. Dr Manuel Liebeke, head of the Metabolomics Department at the CAU’s Faculty of Agricultural and Nutritional Sciences and the Metabolic Interactions Research Group at the Max Planck Institute for Marine Microbiology, a research team has made a breakthrough that provides insights into this mysterious micro-world.
Often, bacteria cannot be cultured in the laboratory, and researchers must rely on information from the bacterial genome obtained from environmental samples to gain theoretical insights into the metabolism of microorganisms. However, there has been a lack of insight into what they actually do in their natural environment. To solve this puzzle, scientists began researching the so-called metabolome of bacteria – everything that has to do with their metabolism, including metabolites such as sugars or fats.
Symbiotic subtenants
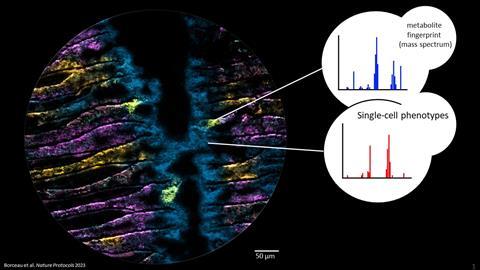
In a pioneering study, Liebeke’s team developed a method with which they can identify individual bacteria and simultaneously determine which metabolites are present in the cells - all without cultivating the bacteria in the laboratory. This method allows them to study how bacteria live and survive as symbiotic subtenants, for example in mussels. The team analysed hundreds of metabolites on an area smaller than one square millimetre. The Kiel and Bremen researchers published their results in the journal Nature Protocols in September.
“We create a snapshot, so to speak, of the bacteria at work, exactly as they are active in their natural environment, particularly within an animal cell,” Liebeke explained. “And we can do that at an impressive resolution of a few micrometres, about ten times thinner than a human hair.”
Flash-frozen tissue
A special feature of this method is the use of flash-frozen tissue, which is cut wafer-thin. The researchers then use a special mass spectrometry technique called MALDI-MS imaging to create a snapshot of the chemical compounds in the cells.
However, drawing the correct conclusions from the images of the metabolites is only possible if they know which bacteria produce or use them. To solve this problem, the researchers also use fluorescence in situ hybridisation (FISH) to identify and localise individual bacterial cells in the sample.
“Applying this method to host-microbe communities will give us many exciting new insights into chemical communication between organisms,” said Patric Bourceau from the Max Planck Institute for Marine Microbiology, lead author of the protocol developed to apply the method.
Opening new doors
This groundbreaking work opens new doors for the study of bacteria and their interactions with their host. In addition, the method presented here also offers promising potential applications for the future: developed at the Max Planck Institute in Bremen, Liebeke’s new working group at the CAU is now using it to study the human gut microbiome and its influence on metabolism. This could, for example, help us to better understand inflammatory bowel diseases. With the publication of a detailed protocol, the application of the technique is now open to other researchers worldwide.
In summary, the application of microscopy and metabolomics (the name of the research field dedicated to the study of metabolites) provides insights into the functional and chemical ecology of host-microbe interactions. The steady advances in MALDI-MSI technology make it possible to illustrate microbial colonies, biofilms and individual eukaryotic cells, and even bacterial microcolonies. Today, MALDI-MSI technology is on the verge of being able to provide images of individual bacterial cells. The protocol presented here forms the basis for analysing and understanding metabolic interactions, down to the micrometre.






No comments yet